Chapter 7 Comparing many groups, interactions, and posterior predictive checks
We’ve been making our models more and more complicated, but we’ve still only fit models with extremely simple fixed effects structures. For example, in the last two chapters we talked about comparing only two groups. Although the comparison of two groups is very simple, it also comes up often. In addition, more complicated problems are often broken down into several two-group questions in the course of understanding a data set. However, real-world experiments don’t usually begin as two-group questions. In this chapter, we’re going to talk about the comparison of observations across multiple groups and using multiple factors.
7.1 Chapter pre-cap
In this chapter, we introduce models that include factors with many levels, and multiple different factors. After that, we discuss the concepts of within and between-subjects factors with respect to factorial designs, orthogonality, and interactions. We then introduce posterior prediction, and the use of this for model checking. Interactions and interaction plots are discussed, as is the way that these can be used to understand main effects and simple main effects. A model with two factors and an interaction, and random effects for all predictors, is fit, and the model is discussed and interpreted. We then present Bayesian \(R^2\) as a simple measure of model fit, useful for getting an indication of how much two models actually differ. Finally, we discuss type S and type M errors, regions of practical equivalence, and the problem of how to know when effects are ‘real’.
7.2 Comparing four (or any number of) groups
First, we’re going to discuss models that compare observations across any number of groups (i.e., levels of a single factor). A model that can be used to compare multiple groups might have just one predictor factor, but that factor can have any number of levels. Although it’s not very common to have a study design or model that only consists of a single multi-group comparison, the inclusion of multi-group comparisons is ubiquitous in many disciplines (including linguistics). Examples of predictors with multiple levels are language (e.g., participants’ first languages), word category (e.g., part of speech, or usage frequency bins), and age groups (e.g., infant, child, teen, adult).
In figure 7.1 we see two ways that subjects (or listeners, or the levels of any grouping factor) can be arranged into groups. In the within-subjects version of this design, each subject is tested at each level of \(A\). In the between-subjects version of the design, different subjects are tested at every level of \(A\). Here we omit the more complicated design discussed in chapter 5, in which some subjects appear at all levels of \(A\) and others don’t. Such designs are definitely analyzable with brms, and sometimes they may be unavoidable. However, they are not included in figure 7.1 because there are a large number of possible configurations of this, and because the additional complexity of such designs can make them extremely challenging to use effectively, so we are typically better off not using such designs if at all possible.

Figure 7.1: Data from a single group of subjects (S) divided according to a single grouping factor A.
7.2.1 Data and research questions
Below we load the data for our experiment investigating apparent speaker height, in addition to the brms and bmmb packages. For the first time, we’re going to analyze productions from all speaker categories at the same time (i.e. boys, girls, men, and women).
library (brms)
library (bmmb)
options (contrasts = c('contr.sum','contr.sum'))
data (exp_data)In figure 7.2 we see that the distribution of apparent height with respect to apparent speaker category is very similar to its distribution with respect to veridical speaker category.
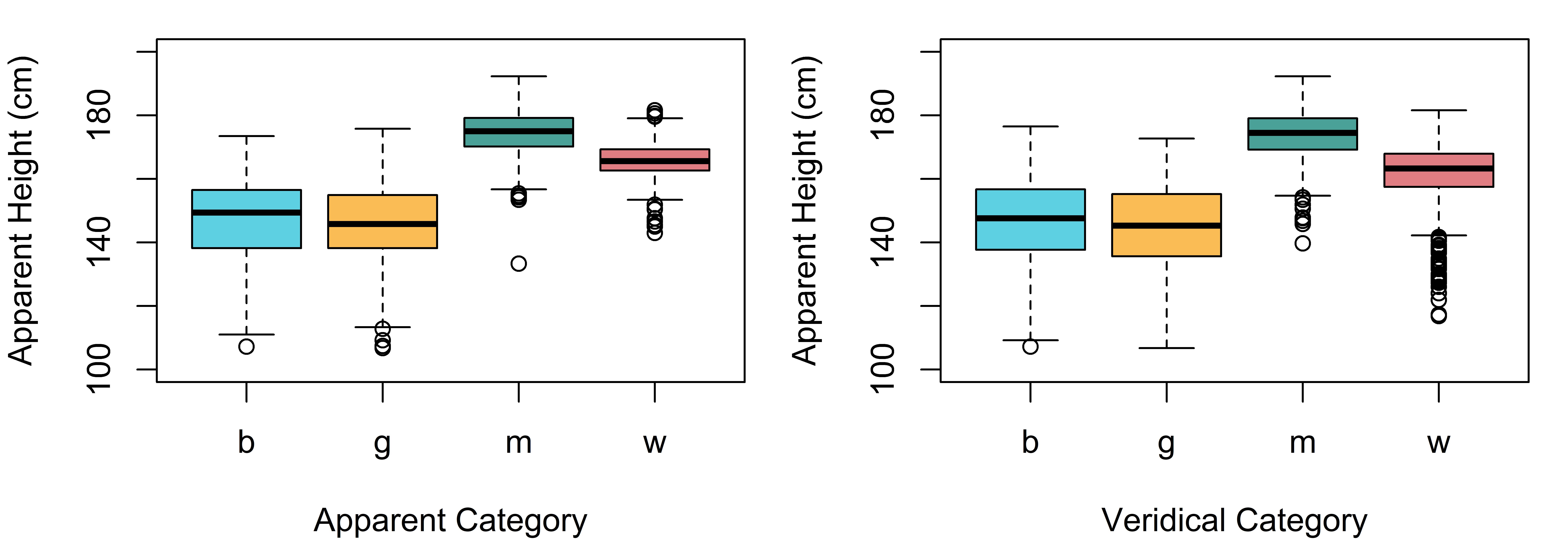
Figure 7.2: Distribution of apparent height judgments for boys (b), girls (g), men (m), and women (w) based on apparent speaker category (left) and veridical speaker category (right).
This is not terribly surprising since as we can see below, listeners identified speaker category correctly in 75% of cases. In addition, errors were most common for boys and girls, speakers that are expected to be approximately the same size.
# overall average correct category identification
mean(exp_data$C == exp_data$C_v)
## [1] 0.7458
# average correct category identification by category
tapply(exp_data$C == exp_data$C_v, exp_data$C_v, mean)
## b g m w
## 0.5778 0.6456 0.9274 0.7097In this chapter we’re going to focus on the effect of apparent speaker category on apparent height. This is because we want to understand how listener’s assumptions about the speaker affect their height judgments, and listeners did not know the veridical speaker category. We’re first going to treat the four groups as if they had no internal relationships between the groups. This means we will be ignoring the fact that we might expect some degree of similarity in the relationships between women and girls, on the one hand, and men and boys, on the other. This may not be the best approach for this data since we know there are logical ways to group boys, girls, men, and women. However, this is a good starting point since in many cases you will have levels of a factor with no logical internal divisions.
Even with only four different groups our potential research questions are substantially more complicated than in the two-group case. First, there are four groups now, meaning we could potentially make 6 2-group comparisons. Second, the groups differ along multiple dimensions, making it more difficult to make two-group comparisons that answer a single question. For example, the ‘man’ and ‘girl’ groups differ according to apparent adultness and apparent gender. In the presence of a difference between groups, this makes it difficult to isolate the contribution of each characteristic to the difference in apparent height. For now, we’re going to keep things simple and just build a model that can tell us about our average group values.
To analyze data like this it should be in a data frame with one row for each observation. One column should contain the dependent variable, the variable whose values and variation you’re trying to predict. Another column should contain the fixed effect predictor determining which group each observation belongs to. Finally, you need one or more columns indicating the sources of data in your experiment. The model in this section will use the following variables from our data frame:
L: A number from 1-15 indicating which listener responded to the trial.C: A letter representing the speaker category (b=boy,g=girl,m=man,w=woman) reported by the listener for each trial.height: A number representing the height (in centimeters) reported for the speaker on each trial.S: A number from 1-139 indicating which speaker produced the trial stimulus.
And we are going to ask the following basic question:
(Q1) Does apparent speaker height vary systematically across apparent speaker categories?
7.2.2 Description of our model
In the last two chapters we discussed the inclusion of a single factor with two levels in our models. Our model formula does not change in structure to include predictors with many levels. The formula to fit our model will be:
height ~ C + (C|L) + (1|S)
Where C is the apparent speaker category indicated by the listener for each trial, a factor with four levels (see above). Since we include C on the left-hand-side of the pipe of L, we know that we are estimating the category by listener interaction using adaptive partial pooling. At this point we have to make a decision regarding how we represent our models in more formal notation, and striking a balance between readability and models that are directly analogous to the output in R.
Each level of the category predictor that we are estimating needs to be represented by a model parameter. Since we are using sum coding for all our factors (see section 5.6.2), our model will estimate three of the four group effects, omitting the alphabetically last level (in this case ‘woman’). As a result, the formula above is analogous to:
height ~ C1 + C2 + C3 + (C1 + C2 + C3|L) + (1|S)
Where C1, C2, and C3 are parameters associated with the first three levels of apparent speaker category. In section 4.3, we noted that each parameter in our model needs to be associated with a predictor. If we call the predictors x1, x2, and x3 our model formula could sort of be thought of as seen below. Of course this is no longer a real or working model formula, we’re just trying to show how \(C\) might actually be included in our model.
height ~ x1*C1 + x2*C2 + x3*C3 + (x1*C1 + x2*C2 + x3*C3|L) + (1|S)
When a factor has only two levels, the effect for one level must equal the negative of the effect for the other level. As a result, group membership for observations can be represented using a single predictor equal to either -1 or 1 (for sum coding, discussed in section 5.7.1). When the observation is associated with one level of the factor the predictor is 1, and when the observation is associated with the other level the predictor is -1. In that way, a single predictor and coefficient can reflect two different levels of the factor.
When there are more than two levels, each level that is being estimated is associated with a predictor variable, meaning that we expect \(J-1\) estimated coefficients for a factor with \(J\) levels in the absence of additional constraints. An example of this is seen below, where we generate three ‘fake’ predictors to represent four groups. Imagine the tiny matrix below represents the numerical predictors for our C parameters so that the first, second, and third columns represent x1, x2 and x3 respectively. Each row is represented by a single 1 save for the final row which is represented by -1 in all other predictors for that factor.
contr.sum(1:4)
## [,1] [,2] [,3]
## 1 1 0 0
## 2 0 1 0
## 3 0 0 1
## 4 -1 -1 -1Our tiny data matrix above suggests that the first observation (row) belongs to the first group (boy). This is because our data results in the \(C1\) parameter being multiplied by one, and \(C2\) and \(C3\) being zeroed out (see section 4.3). The same logic suggests the second row is an observation from the second group and the third from the third. For example, in (7.1) we imagine that the first four values of our C predictor are 1, 2, 3, and 4, meaning that the apparent speaker category indicated for the first four trials were boy, girl, man, and woman. We also imagine that our predicted value (\(\mu\)) is a combination of our estimated parameters \(C1, C2\) and \(C3\) with the ‘secret’ predictors in our matrix above. We can see that multiplying our coefficients with our predictors has the effect of ‘erasing’ the unwanted coefficients for a given row leaving only the one associated with the observation.
\[ \begin{equation} \begin{split} \mathsf{C}_{[1]} = 1, \mathsf{C}_{[2]}=2,\mathsf{C}_{[3]}=3,\mathsf{C}_{[4]}=4 \dots \\ \mu_{[i]} = x_1 \cdot C1 + x_2 \cdot C2 + x_3 \cdot C3 \\ \\ \mu_{[1]} = 1 \cdot C1 + 0 \cdot C2 + 0 \cdot C3 = C1\\ \mu_{[2]} = 0 \cdot C1 + 1 \cdot C2 + 0 \cdot C3 = C2\\ \mu_{[3]} = 0 \cdot C1 + 0 \cdot C2 + 1 \cdot C3 = C3\\ \mu_{[4]} = -1 \cdot C1 - 1 \cdot C2 - 1 \cdot C3 = -(C1 + C2 + C3) = C4\\ \end{split} \tag{7.1} \end{equation} \]
Since \(C4\) is equal to \(-(C1+C2+C3)\) (under sum coding), adding the negative of \(C1\), \(C2\), and \(C3\) to the prediction is equivalent to adding the value of \(C4\). As a result, the fourth row of our tiny matrix represents an observation from the fourth group, and encodes the effect for \(C4\), even without directly estimating this value. As a result, even though our model only directly estimates three parameters for our \(C\) predictor, we could think of our model as tacitly representing four parameters for this factor: \(C1\), \(C2\), \(C3\), and \(-(C1+C2+C3)\).
Even though the inclusion of \(C\) in our model requires the addition of three separate parameters, we’re going to represent these with a single term like \(C_{[\mathsf{C}_{[i]}]}\) in our formal model descriptions. This term selects the value of \(C\) for trial \(i\) based on the value of the C (apparent category) predictor, which is consistent with the way we’ve been representing our listener and speaker-dependent intercepts. However, there appears to be a problem. The subscript of the term is a predictor which has four levels (C, or \(\mathsf{C}\)). When the C predictor is equal to it’s fourth level (C = w), the model seems to want to select the fourth \(C\) predictor, i.e. \(C_{[4]}\). However, our \(C\) predictor vector does not contain four elements because the fourth level of the C predictor is not estimated (i.e. there is no \(x_4\) predictor in (7.1)). So, when this occurs we can just imagine that our model reconstructs the missing parameter using the ones that are estimated, and then uses this value for \(C4\).
Since we are including adult male speakers in our data we should also reconsider our priors. Based on the information in height_data we expect average apparent heights of around 176, 162, 150 and 150 cm respectively for men, women, girls, and boys. These four groups have an average of 159 cm and a standard deviation of 12 cm. Based on this, and especially given that we are investigating apparent and not veridical height, the priors we used in the last chapter still seem appropriate. The formal description of our model is given below. We omit the deterministic equation building up \(\mathrm{\Sigma}\) from the correlation matrix (\(R\)) and the individual standard deviations (shown in (6.11)) since this does not involve the user, and it makes the model description needlessly bigger.
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{t}(\nu, \mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + C_{[\mathsf{C}_{[i]}]} + L_{[\mathsf{L}_{[i]}]} + C_{[\mathsf{C}_{[i]}]} \colon L_{[\mathsf{L}_{[i]}]} + S_{[\mathsf{S}_{[i]}]} \\ \\ \mathrm{Priors:} \\ S_{[\bullet]} \sim \mathrm{N}(0,\sigma_S) \\ \begin{bmatrix} L_{[\bullet]} \\ C_{[1]} \colon L_{[\bullet]} \\ C_{[2]} \colon L_{[\bullet]} \\ C_{[3]} \colon L_{[\bullet]} \end{bmatrix} \sim \mathrm{MVNormal} \left(\begin{bmatrix} 0 \\ 0 \\ 0 \\ 0 \\ \end{bmatrix}, \mathrm{\Sigma} \right) \\ \\ \mathrm{Intercept} \sim \mathrm{N}(156,12) \\ C_{[\bullet]} \sim \mathrm{N}(0,12) \\ \sigma_L,\sigma_{C_{[1]} \colon L},\sigma_{C_{[2]} \colon L},\sigma_{C_{[3]} \colon L}, \sigma_S \sim \mathrm{N}(0,12) \\ \nu \sim \mathrm{gamma}(2, 0.1) \\ R \sim \mathrm{LKJCorr} (2) \end{split} \tag{7.2} \end{equation} \]
Structurally, this model is extremely similar to the one described in (6.11) last chapter with \(C_{[\mathsf{C}_{[i]}]}\) replacing \(A\) and \(C_{[\mathsf{C}_{[i]}]} \colon L_{[\mathsf{L}_{[i]}]}\) replacing \(A \colon L_{[\mathsf{L}_{[i]}]}\). Note that since we are estimating three parameters for C, our listener-dependent ‘random effects’ come from a four-dimensional normal distribution which considers the relationships between the intercepts and category effects for individual listeners. Here is a plain English description of the model in (7.2):
We are modeling apparent height as coming from a t distribution with unknown nu (\(\nu\)), mean (\(\mu\)), and scale (\(\sigma\)) parameters. The expected value for any given trial (\(\mu\)) is modeled as the sum of an intercept, an effect for apparent speaker category (\(C\)), a listener effect (\(L\)), listener dependent effects for apparent category (e.g. \(C_{[1]} \colon L\)), and a speaker effect (\(S\)). The speaker effects were drawn from a univariate normal distribution with a standard deviation (\(\sigma_S\)) estimated from the data. The four listener effects were drawn from a multivariate normal distribution with individual standard deviations (\(\sigma_L,\sigma_{C_{[1]} \colon L},\sigma_{C_{[2]} \colon L},\sigma_{C_{[3]} \colon L}\)) and a correlation matrix (\(R\)) that was estimated from the data. The remainder of the ‘fixed’ effects and correlations were given prior distributions appropriate for their expected range of values.
7.2.3 Fitting and interpreting the model
We fit the four-group model below:
# Fit the model yourself
priors = c(brms::set_prior("student_t(3,156, 12)", class = "Intercept"),
brms::set_prior("student_t(3,0, 12)", class = "b"),
brms::set_prior("student_t(3,0, 12)", class = "sd"),
brms::set_prior("lkj_corr_cholesky (2)", class = "cor"),
brms::set_prior("gamma(2, 0.1)", class = "nu"),
brms::set_prior("student_t(3,0, 12)", class = "sigma"))
model_four_groups =
brms::brm (height ~ C + (C|L) + (1|S), data = exp_data, chains = 4,
cores = 4, warmup = 1000, iter = 5000, thin = 4,
prior = priors, family = "student")# or download it from the GitHub page:
model_four_groups = bmmb::get_model ('7_model_four_groups.RDS')We’ll focus on the fixed effects, seen below:
brms::fixef(model_four_groups)
## Estimate Est.Error Q2.5 Q97.5
## Intercept 158.581 1.083 156.41 160.685
## C1 -9.318 1.310 -11.87 -6.682
## C2 -12.082 1.494 -15.07 -9.110
## C3 15.286 1.327 12.59 17.950It’s always a good idea to make sure your model coefficients make sense given your data. Doing things like this will help minimize mistakes and make sure that you really understand your model. We can see that the intercept corresponds to the average of the group means, and our C coefficients correspond reasonably to, but don’t exactly match, the centered group means.
# group means
means = tapply (exp_data$height, exp_data$C, mean)
# Intercept = mean of means
mean (means)
## [1] 158
# Group effects = centered group means
means - mean (means)
## b g m w
## -11.12 -13.03 16.56 7.59There are many possible reasons for this including the fact that the group means don’t account for repeated measures, a lack of balance in the data, or for the way that outliers can unduly influence sample mean estimates. That being said, it’s useful to see that our model coefficients seem to reflect the data and we should be concerned if these diverged wildly. Note that, as discussed above, we’re missing the C4 coefficient. We can use the short_hypothesis function to easily recover the final group coefficient by finding the negative sum of the coefficients that were estimated. An example of this is shown below.
# missing group effect
bmmb::short_hypothesis (model_four_groups, c("-(C1+C2+C3) = 0"))
## Estimate Est.Error Q2.5 Q97.5 hypothesis
## H1 6.114 1.216 3.686 8.543 (-(C1+C2+C3)) = 0Clearly, there are differences in apparent height across groups. If someone asked you “please describe the differences to me”, you might begin by saying something like “there doesn’t seem to be too much difference in apparent height between apparent boys and girls”, or “apparent men are identified as taller than apparent women”. In each case, these statements reduce a multi-group model to focus on a single difference between groups at a time. We can use short_hypothesis to help us compare different combinations of our fixed effects. Using the code below we test for a difference in group means between the first and second groups (boys and girls), and between the third and fourth groups (men and women).
# find differences between groups
comparisons = bmmb::short_hypothesis (model_four_groups,
c("C1 = C2", "C3 = -(C1+C2+C3)"))We can use brmplot from the bmmb package to easily make plots showing means and credible intervals for our fixed effects, including the recovered coefficient. We also plot the two comparisons we made using the hypothesis function above, presented in figure 7.3.
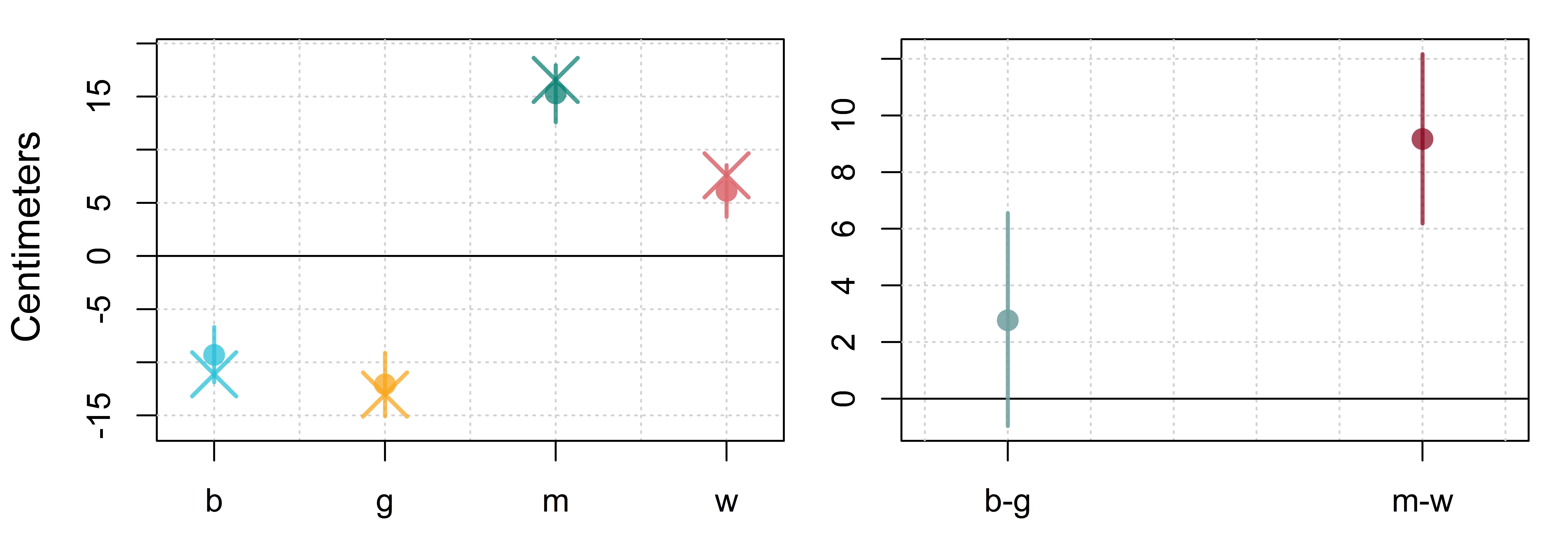
Figure 7.3: (left) Means and 95% credible intervals for boy (b), girl (g), man (m), and woman (w) effects for our four group model. (right) Means and 95% credible intervals for differences between boys and girls, and men and women.
The distributions in the left plot of the figure tell us about probable values for our category parameters: Given our data and model structure, there is a 95% probability that the parameter value falls within this interval. Our category effects reflect the expected difference between our model intercept, the overall grand mean, and each category mean. So, the intervals on the left of figure 7.3 tell us that there is a 95% probability (given our data and model structure) that the actual difference between the grand mean and each group mean falls within that range. What do the distributions in the right plot in figure 7.2 tell us? They represent the posterior distribution of the difference between category means directly, without involving the intercept.
Conceptually, finding the difference between the intercept and a group mean is no different than finding the difference between individual group means. Consider the example of providing directions with respect to your house. If the library is 5 miles west and the park is 3 miles west, you could refer to these as -5 and -3 miles from your house, west to east. However, you could also express the difference between one and the other directly as 2 (or -2). The left plot in figure 7.2 expresses all distances with respect to the intercept (your house) while the right plot expresses differences with respect to individual category means directly (the library and park).
7.3 Investigating multiple factors simultaneously
Our model above had a single factor with four groups/levels. Another way to look at it is that the model predicted variation along a single dimension, that is a single piece of information that can be used to distinguish our groups. However, we can usually categorize people (or observations) along multiple dimensions simultaneously - there is more structure in the data than a single dimension captures. So, a speaker may be an adult and female or adult and male, child and female or child and male, and not just adult or child, male or female. This is because adultness and gender are separate characteristics (dimensions) and the value of one does not determine the value of the other. In our height experiment we effectively asked listeners to make two binary classifications: Is the listener an adult or a child? Is the listener female or male? This means we can think of our four groups as varying along two dimensions, each represented by a different factor. However, although we know this, our previous model didn’t: It thinks we just have four groups that have no logical internal distinctions.
To include multiple predictors in our model and inspect the effect of all combinations of factor levels on our dependent variable, we must cross our predictors. This means that we observe each level of one factor at every level of the other factor (and vice versa). For our data this means that we observe both male and female children, and male and female adults. Crossing factors results in \(n_A \cdot n_B\) individual groups for two factors (\(A\) and \(B\)) with \(n_A\) and \(n_B\) number of levels each. Since we have two factors with two levels each, crossing apparent age and gender results in four unique groups: Boys (male child), girls (female child), men (male adult), and women (female adult). Designs where all the factors are crossed are sometimes called factorial or orthogonal designs.
When you have orthogonal designs, that means that the effect of your ‘dimensions’ (i.e., factors) can be estimated separately. For example, if you want to know the difference in apparent height across levels of apparent gender, it helps to have observed both adults and children for each gender. A comparison of boys and women will conflate gender and age differences, thereby making it difficult to understand the independent effect of apparent gender. By balancing factors across each other, orthogonal designs allow us to decompose the variation in our data using different factors more effectively. This allows us to attribute the variation in our observations to the different factors in our design (we will return to this idea in chapter 11).
Figure 7.4 presents a few ways that subjects can be combined with two two-level factors, \(A\) and \(B\), whose combination can represent four groups. First, we can see that factors \(A\) and \(B\) are crossed with each other since there are subjects, and presumably observations, in each little cell (i.e. combination of factor levels). What would it look like for these factors to not be crossed? Well, one of the ‘cells’ (boxes) would be completely missing observations. If this were to happen, we would obviously lose the ability to make comparisons across groups involving the values in that cell. If your subjects factor is fully crossed with your fixed effects factors (in this case \(A\) and \(B\)) then you have a fully within-subjects design. If you only observe certain subjects for specific combinations of fixed effects, you have a fully between-subjects design.

Figure 7.4: Data from groups of subjects (S), divided according to two grouping factors A and B.
Finally, we see a new possibility, a mixed design. In a mixed design, one or more factors are between-subjects, and the others are within-subjects. In the example below, \(A\) is between-subjects but \(B\) is within-subjects. This means that each level of \(A\) has different subjects but each level of \(B\) has the same subjects. An example of a mixed design would arise if, for example, we wanted to run our coffee and reading time experiment (discussed in chapter 5) on multiple first languages. Each subject could be tested at both coffee and decaf, making that factor within subjects, however, each subject would only be observed within a single first language group, making that factor between subjects.
Our model contains two predictors with two levels each, leading to four groups. Height judgments for these four groups are presented in figure 7.5. These four boxes correspond to the four cells in figure 7.4. The middle plot in 7.5 compares height judgments across levels of gender (male vs. female). Since we have both adults and children within each gender group, each box in the plot is really the average of two of the boxes from the left plot in 7.5 (boxes 1 and 3 vs. 2 and 4). This is equivalent to finding the average of one row in 7.4, and comparing it to the average of the other row.
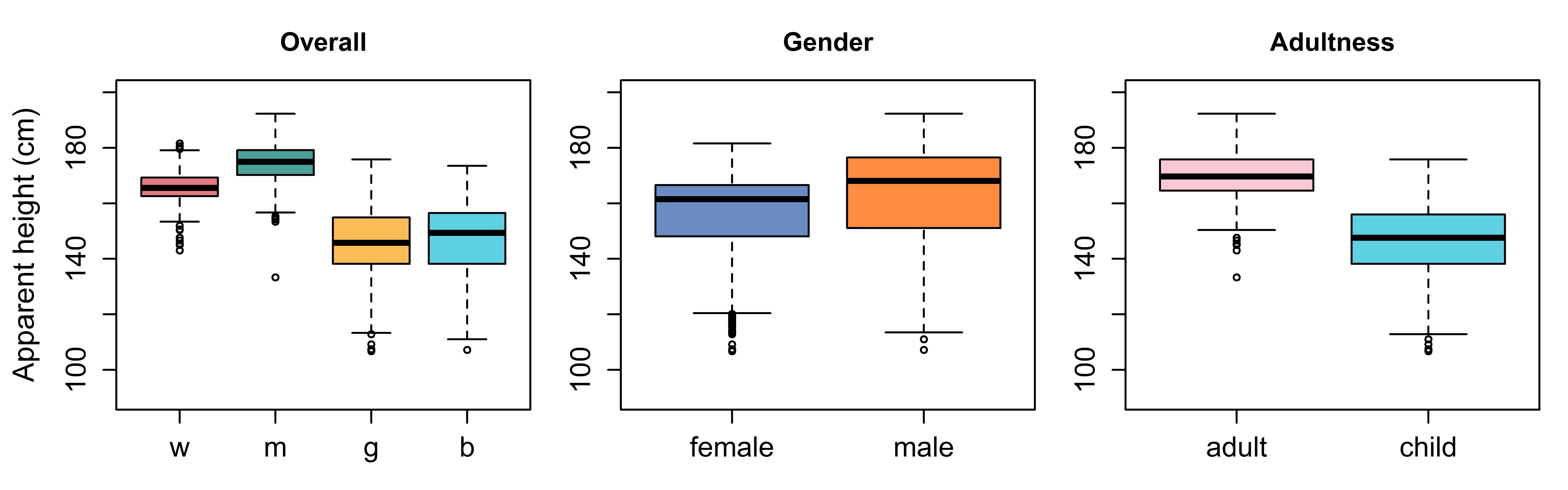
Figure 7.5: (left) Distribution of apparent height judgments for apparent boys (b), girls (g), men (m), and women (w). (middle) Comparison of distributions based on whether the speaker is female or male. (right) Comparison distributions based on whether the speaker is an adult or child.
Similarly, the right plot in 7.4 compares height judgments across levels of age (adult vs. child). Since we have both males and females within each gender group, each box in the plot is really the average of two of the boxes from the left plot in 7.5 (boxes 1 and 2 vs. 3 and 4). This is equivalent to finding the average of one column in 7.4, and comparing it to the average of the other column. In this way, our model including two fixed effect predictors can use four groups to answer two questions simultaneously: What is the effect of apparent age on apparent height? What is the effect of apparent gender on apparent height?
7.3.1 Data and research questions
We load our data and packages again just in case:
library (brms)
library (bmmb)
options (contrasts = c('contr.sum','contr.sum'))
data (exp_data)We’re going to use slightly different set of columns from our data this time, this time using A and G rather than C. The model in this section will use the following variables from our data frame:
L: A number from 1-15 indicating which listener responded to the trial.height: A number representing the height (in centimeters) reported for the speaker on each trial.S: A number from 1-139 indicating which speaker produced the trial stimulus.G: The apparent gender of the speaker indicated by the listener,f(female) orm(male).A: The apparent age of the speaker indicated by the listener,a(adult) orc(child).
And we’re going to split the question we posed above into two questions:
(Q1) Does average apparent height differ across levels of apparent age?
(Q2) Does average apparent height differ across levels of apparent gender?
7.3.2 Description of the model
In our previous model, we acted like we just had four different groups with no internal structure. Of course, we know that our groups differ systematically from each other in meaningful ways. For example, we might have chosen to fit two separate models investigating age and gender like this:
brm (height ~ A + (A|L) + (1|S)
brm (height ~ G + (G|L) + (1|S)
For several reasons (some of which we’ll see very soon), it’s preferable to fit a single model with both predictors at once, rather than fitting separate models for each one. Our model formula will now look like this, reflecting the influence of both predictors simultaneously:
height ~ A + G + (A + G|L) + (1|S)
This can be read like “apparent height is distributed according to effects for apparent adultness and gender, with random intercepts, age, and gender effects for each listener, and random intercepts for each speaker”. You may have noticed that our model no longer includes the C predictor. This is because the C label is perfectly predictable on the basis of A and G (i.e., as a female child, a member of the g group must have values of f and c for their G and A predictors). Basically, we have decomposed the four categories into two components to help us understand the effect of each. For example, the fixed effect predictors in our model were previously:
\[ \begin{equation} \begin{split} \mu_{[i]} = \mathrm{Intercept} + (C_{[\mathrm{C}_{[i]}]}) \end{split} \tag{7.3} \end{equation} \]
We can expand the term in parentheses as seen below. Since group can be exactly represented by combinations of gender and adult, our model sort of ‘secretly’ contained this more-complicated model inside of it.
\[ \begin{equation} \begin{split} \mu_{[i]} = \mathrm{Intercept} + (A_{[\mathrm{A}_{[i]}]} + G_{[\mathrm{G}_{[i]}]}) \end{split} \tag{7.4} \end{equation} \]
The above can be thought of as a ‘decomposition’ of the variance in your model by breaking it up into parts and assigning them to different sources. We have actually been doing this from the start by making more and more complicated models. Recall that our very first approach (in chapter 3) to understanding variation in apparent height treated all variation as error, and so did not ‘decompose’ anything. In chapter 4 we added between-listener and speaker variation to the model, and in chapter 5 we added apparent age. Now we include apparent age and gender, listener specific effects and intercepts, and speaker specific intercepts. There is a fixed amount of variation in our dependent variable. Every tiny bit of variance that these added components explain takes away from the data-level error variance (\(\sigma\)), and potentially also from the other components that had previously been in the model. Our updated model description is provided in (7.5), again omitting the deterministic equation ‘building’ up \(\mathrm{\Sigma}\).
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{t}(\nu, \mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + A + G + L_{[\mathsf{L}_{[i]}]} + A \colon L_{[\mathsf{L}_{[i]}]} + G \colon L_{[\mathsf{L}_{[i]}]} + S_{[\mathsf{S}_{[i]}]} \\ \\ \mathrm{Priors:} \\ S_{[\bullet]} \sim \mathrm{N}(0,\sigma_S) \\ \begin{bmatrix} L_{[\bullet]} \\ A \colon L_{[\bullet]} \\ G \colon L_{[\bullet]} \end{bmatrix} \sim \mathrm{MVNormal} \left( \begin{bmatrix} 0 \\ 0 \\ 0 \\ \end{bmatrix}, \mathrm{\Sigma} \right) \\ \\ \mathrm{Intercept} \sim \mathrm{N}(156,12) \\ A, G \sim \mathrm{N}(0,12) \\ \sigma_L, \sigma_{A \colon L}, \sigma_{G \colon L}, \sigma_S \sim \mathrm{N}(0,12) \\ \sigma \sim \mathrm{N}(0,12) \\ \nu \sim \mathrm{gamma}(2, 0.1) \\ R \sim \mathrm{LKJCorr} (2) \end{split} \tag{7.5} \end{equation} \]
Our models have increasingly large numbers of priors, and many of these are the same. As a result, going forward we will place these on the same line when they have the same prior and represent the same sorts of parameters (in much the same way they are specified by the class parameter in the set_prior function). For example, the line \(\sigma_L, \sigma_{A \colon L}, \sigma_{G \colon L}, \sigma_S \sim \mathrm{N}(0,12)\) specifies the prior probability of all standard deviation terms individually, but at the same time. In plain English this says:
We are modeling apparent height as coming from a t distribution with unknown nu (\(\nu\)), mean (\(\mu\)), and scale (\(\sigma\)) parameters. The expected value for any given trial (\(\mu\)) is modeled as the sum of an intercept, an effect for apparent speaker age (\(A\)) and gender (\(G\)), a listener effect (\(L\)), a listener dependent effect for apparent age (\(A \colon L\)) and apparent gender (\(G \colon L\)), and a speaker effect (\(S\)). The speaker effects were drawn from a univariate normal distribution with a standard deviation (\(\sigma_S\)) estimated from the data. The three listener effects were drawn from a multivariate normal distribution with individual standard deviations (\(\sigma_L,\sigma_{A \colon L},\sigma_{G \colon L}\)) and a correlation matrix (\(R\)) that was estimated from the data. The remainder of the ‘fixed’ effects and correlations were given prior distributions appropriate for their expected range of values.
7.3.3 Fitting and interpreting the model
Below we fit a model with the structure outlined in (7.5). We don’t need to change our priors because our new predictors are all covered by the classes of priors we have specified so far, and we think the priors we have been using thus far are still a good fit for our data.
# Fit the model yourself
priors = c(brms::set_prior("student_t(3,156, 12)", class = "Intercept"),
brms::set_prior("student_t(3,0, 12)", class = "b"),
brms::set_prior("student_t(3,0, 12)", class = "sd"),
brms::set_prior("lkj_corr_cholesky (2)", class = "cor"),
brms::set_prior("gamma(2, 0.1)", class = "nu"),
brms::set_prior("student_t(3,0, 12)", class = "sigma"))
model_both =
brms::brm (height ~ A + G + (A + G|L) + (1|S), data = exp_data,
chains = 4, cores = 4, warmup = 1000, iter = 5000,
thin = 4, prior = priors, family = "student")# Or download it from the GitHub page:
model_both = bmmb::get_model ('7_model_both.RDS')We can inspect the model fixed effects:
# inspect the fixed effects
brms::fixef (model_both)
## Estimate Est.Error Q2.5 Q97.5
## Intercept 158.654 1.1947 156.319 161.025
## A1 9.979 1.2107 7.562 12.282
## G1 -2.205 0.5524 -3.288 -1.124We now have two non-Intercept ‘Population-Level’ effects: A1 and G1, representing the categories ‘adult’ and ‘female’ respectively. Remember that since we used sum coding and have only two levels for each factor, the effects for the groups that are not represented (‘child’, ‘male’) are just the opposite sign of the groups that are represented. Below, we calculate the mean apparent height of the four groups and find the grand mean of the four groups. We can see that this is very similar to our intercept.
# Intercept
mean (tapply (exp_data$height, exp_data$C, mean))
## [1] 158Next we find the average apparent height for each level of adultness, find the difference between these, and divide it by negative two (just to flip the sign to match our effects). We also do the same for the levels of age. This tells us half the difference between our group means, and we can see that these values are reasonably similar to the estimated A1 and G1 predictors.
# Age effect
diff (tapply (exp_data$height, exp_data$A, mean) ) / -2
## c
## 12.16
# Gender effect
diff (tapply (exp_data$height, exp_data$G, mean) ) / -2
## m
## -3.166We can recover the four original group means by adding up individual fixed effect coefficients. This can sometimes be tedious and requires you to be careful and methodical, but isn’t actually difficult. Remember that each of the four groups is uniquely identified by a combination of gender and adultness. This means that to recover the expected group means, we need to add the right combination of coefficients to the intercept. For example, the first hypothesis we test below (Intercept + -A1 + -G1 = 0) takes the intercept and adds the effect for ‘child’ (-A1). We use -A1 and not A2 (i.e. the second level of A) because our model does not contain a parameter called A2. This parameter is not estimated in our model because -A1=A2. So, if we want the value of A2 we use -A1. Similarly, we add the effect for ‘male’ using -G1. Since we started with the overall mean apparent height (the intercept) and add the effects for ‘child’ and ‘male’, the hypothesis Intercept + -A1 + -G1 = 0 estimates the group mean for ‘boys’.
means_pred = bmmb::short_hypothesis (model_both,
c("Intercept + -A1 + -G1 = 0", # boys
"Intercept + -A1 + G1 = 0", # girls
"Intercept + A1 + -G1 = 0", # men
"Intercept + A1 + G1 = 0")) # women
means_pred
## Estimate Est.Error Q2.5 Q97.5 hypothesis
## H1 150.9 2.2410 146.6 155.3 (Intercept+-A1+-G1) = 0
## H2 146.5 2.3680 142.0 151.2 (Intercept+-A1+G1) = 0
## H3 170.8 1.2988 168.2 173.4 (Intercept+A1+-G1) = 0
## H4 166.4 0.6901 165.0 167.8 (Intercept+A1+G1) = 0Below we compare our recreated group means with our average height judgment for each group. There is a reasonable similarity, however there also appear to be some important mismatches.
tapply (exp_data$height, exp_data$C, mean)
## b g m w
## 146.9 145.0 174.5 165.6For example, we see that adult women and men differ in height by 9 cm in our data, while boys and girls by only 2 cm. So, it seems like the difference between males and females is smaller for children than for adults. However, according to our model men and women differ by about 4.4 cm, and so do and boys and girls. This discrepancy suggests that our model is potentially missing important information about our data.
7.4 Posterior prediction: Using our models to predict new data
So far we’ve been working with very simple models and so we haven’t worried very much about how well they fit, or represent our data. Our reconstruction of the group means above suggests our current model may have some issues, and that we should be concerned about its ability to accurately capture the patterns in the data. So far we’ve only been discussing modeling in one direction, going from data to abstract, reduced representations (i.e. regression models). However, it’s also worth considering going in the other direction, from the abstract representation back towards data.
One way to see how well our model represents our data is to consider what data would be generated by the relationships represented in our model. Using your model in this way is called prediction, and you can use prediction to evaluate the fit of your model. The reasoning is basically, if your model is a good representation of the data, the ‘fake’ data it generates should look very much like your real data. If the fake data it generates doesn’t look like your real data, something about your model is slightly ‘off’. The discussion of prediction presented here will involve dependent variables that do not require a link function (discussed in chapter 10). Some of the details discussed here, in particular those involving expected values, do not exactly apply to models that require a link function.
All our models so far have contained lines like those seen in (7.6). These lines tell our models that: 1) We expect our data to be normally distributed given some mean and standard deviation, and 2) The mean for a given trial can be predicted using some combination of the independent variables.
\[ \begin{equation} \begin{split} y_{[i]} \sim N (\mu_{[i]}, \sigma) \\ \mu_{[i]} = Intercept + A_{[\mathsf{A}_{[i]}]} + B_{[\mathsf{B}_{[i]}]} + C_{[\mathsf{C}_{[i]}]} \end{split} \tag{7.6} \end{equation} \]
The value of \(\mu\) above is the expected value for a given trial. This is also sometimes called the linear predictor, or the conditional expectation function (in econometrics). Since the linear predictor is the value you expect for a trial, it’s obvious that there should be a reasonable correspondence between the linear predictor and your dependent variable. If what your model tells you to expect is not like what you actually observe, then what good is the model? One shortcoming of the linear predictor is that this does not incorporate the random trial-to-trial error (i.e., \(\sigma\)) that you know is a part of your data. As a result, the linear predictor is constant for any single combination of dependent variables, and undersells the amount of variation that can be expected in a given data set.
The posterior predictive distribution is the distribution of possible data given your parameter estimates and probability model. In other words, the posterior predictive distribution uses your linear predictor, but also includes the data-level noise implied by the structure of your model. For normally distributed data, our posterior predictive distribution is the distribution of generated values of \(\tilde{y}\) given estimates of \(\mu\) and \(\sigma\) as in (7.7). We use \(\tilde{y}\) to represent new observations, as opposed to \(\hat{y}\) which represents estimates of current data.
\[ \begin{equation} \begin{split} \tilde{y}_{[i]} \sim N (\mu_{[i]}, \sigma) \\ \end{split} \tag{7.7} \end{equation} \]
A posterior predictive check samples from the posterior predictive distribution with the intention of comparing these samples of \(\tilde{y}\) to your original data \(y\). Posterior prediction can be used to assess how well your model fits your data. If your model really ‘gets’ your data, the fake data it generates (\(\tilde{y}\) ) will have similar characteristics to your real data (\(y\)). If the fake data your model generates looks substantially different from your real data, that suggests a fundamental misalignment between your model and your data.
You can get simple representations of your posterior predictive distribution using the pp_check function in brms as seen below. This function will compare the distribution of your new data (\(\tilde{y}\)) to the distribution of your actual data. It compares density plots by default, though the user can select other sorts of plots.
brms::pp_check(model_both)You can also use the p_check (predictive check) function in brms to make the same sorts of comparisons, though this function only compares densities. This function lets you hide or show your data in the plot, lets you specify the number of posterior samples you want, and also lets you specify the random effects formula when predicting. This function also returns a matrix containing the posterior predictions (y_pred below) where each row represents a different observation and each column represents a different posterior sample.
y_pred = bmmb::p_check(model_both, show_data = TRUE)The functions above are good for a ‘30,000-foot view’ of our posterior predictions. They show you when something big has gone wrong but may not let you see if something small goes wrong. To investigate nuances in your predictions, you may need to consider these using the same sort of plots you would use to interpret your real data, for example using a representation as seen in figure 7.5. The generic functions fitted and predict will help you get linear and posterior predictions from your brm model.
# linear predictor
y_lin_pred = fitted (model_both)
# posterior prediction
y_post_pred = predict (model_both)In both cases the functions return four-column data frames with as many rows as the number of observations in the data. These data frames contain columns representing the mean, standard deviation and the upper and lower 95% credible intervals for each one of our data points. The reason we get intervals for our posterior predictions is because our model has a different prediction for each set of posterior samples. This means that if we have 4000 samples we actually have 4000 slightly different sets of parameter estimates and so 4000 slightly different predictions. So, in addition to information about the average prediction for each data point, we get information about variation around these estimates. We can see the (summarized) data frame representing our linear predictors.
# linear predictions
head (y_lin_pred)
## Estimate Est.Error Q2.5 Q97.5
## [1,] 156.8 2.182 152.5 161.0
## [2,] 161.7 2.059 157.6 165.6
## [3,] 160.6 1.983 156.6 164.3
## [4,] 162.3 1.846 158.5 165.9
## [5,] 163.0 1.992 159.0 167.0
## [6,] 155.7 2.371 151.1 160.5And below we see the data frame corresponding to our posterior predictions. Note that although the means of the linear and posterior predictions are very similar, the intervals around the posterior predictions are noticeably wider. This is because the posterior predictions incorporate the data-level error (\(\sigma\)) in their predictions. Another difference between the linear and posterior predictions is that the linear predictors are fixed for a given set of posterior samples. In contrast, every time you run the command predict(model) you will get slightly different results because these incorporate random error.
# posterior predictions
head (y_post_pred)
## Estimate Est.Error Q2.5 Q97.5
## [1,] 157.0 7.915 141.3 172.2
## [2,] 161.7 9.137 146.3 177.2
## [3,] 160.5 8.210 144.8 175.9
## [4,] 162.1 8.312 146.4 177.0
## [5,] 162.8 7.639 147.8 177.9
## [6,] 155.7 7.708 140.0 170.8For both kinds of predictions, you can also set summary=FALSE to get the individual (unsummarized) predictions. If you do this you get individual predictions for every set of posterior samples for your parameters, for every data point. So, if you have 1500 data points and 4000 posterior samples, the unsummarized output of these functions would be a matrix with 4000 rows and 1500 columns.
There’s a parameter for the predict function, re_formula, that determines which random effect terms are included in your prediction. By default this parameter is set to NULL meaning that your predictions will be made using your complete model formula. For example for our last model this means our posterior predictions reflect this model formula:
height ~ A + G + (A + G|S) + (1|L)
We can instead set re_formula=NA to make predictions using only the fixed effects as seen below.
y_post_pred_no_re = predict (model_both, re_formula = NA)This corresponds to the predictions made by this model:
height ~ A + G
We could also include a subset of random effects, for example including only the listener random intercepts as seen below:
y_post_pred_some_re = predict (model_both, re_formula = "~(1|L)")This would generate predictions made by the following model:
height ~ A + G + (1|L)
Below we sample from the posterior distribution but ask for only a single (randomly chosen) prediction. We do this for our full model and for a model that makes only fixed effects predictions.
y_hat = predict (model_both, ndraws = 1)
y_hat_no_re = predict (model_both, re_formula = NA, ndraws = 1)In figure 7.6 we compare the distribution of our data to our posterior predictive distributions, without and with the inclusion of random effects.

Figure 7.6: (left) Distribution of apparent height judgments according to apparent speaker category (boy, girl, man, woman) for our real data. (middle) Posterior predictions made by model_both without random effects (RE). (right) Posterior predictions made by model_both with random effects (RE).
There are some differences in the ‘widths’ of the boxes, but we will not discuss this for now. Instead, we would like to highlight differences in the relative positions of the boxes representing the four groups across the plots. Our data shows different average heights for men and women but no noticeable height difference between boys and girls. Despite this, both sets of our posterior predictions show a difference in height between boys and girls. This is particularly evident in our fixed effects predictions since the random effects can help represent many of the idiosyncrasies of any given data set. As we will discuss in the following section, the structure of our data (and our world knowledge) suggests that there is an interaction between apparent gender and apparent age, meaning that we should include the interaction between these predictors in our model.
7.5 Interactions and interaction plots
We can think of a single effect representing a difference between groups/conditions/levels as a slope. For example, in the left plot in figure 7.7 we plot mean apparent height for apparent females and males at arbitrary x-axis locations. The difference in the group means is 6 cm (females 157 cm, males 163 cm). We can use any arbitrary x axis distance to calculate slopes, as long as we are consistent. However, there are obvious practical advantages to choosing to calculate these slopes over the arbitrary ‘distance’ of 1. To do this, we can imagine that we plot the first level (female) at 0 and successive levels at sequential integer values along the x axis (i.e. 1, 2, 3, …). When we do this, the line formed by joining the averages of these groups has a slope of 6 (i.e., it rises 6 cm from 0 to 1, from one group to another, for a slope of 6/1).
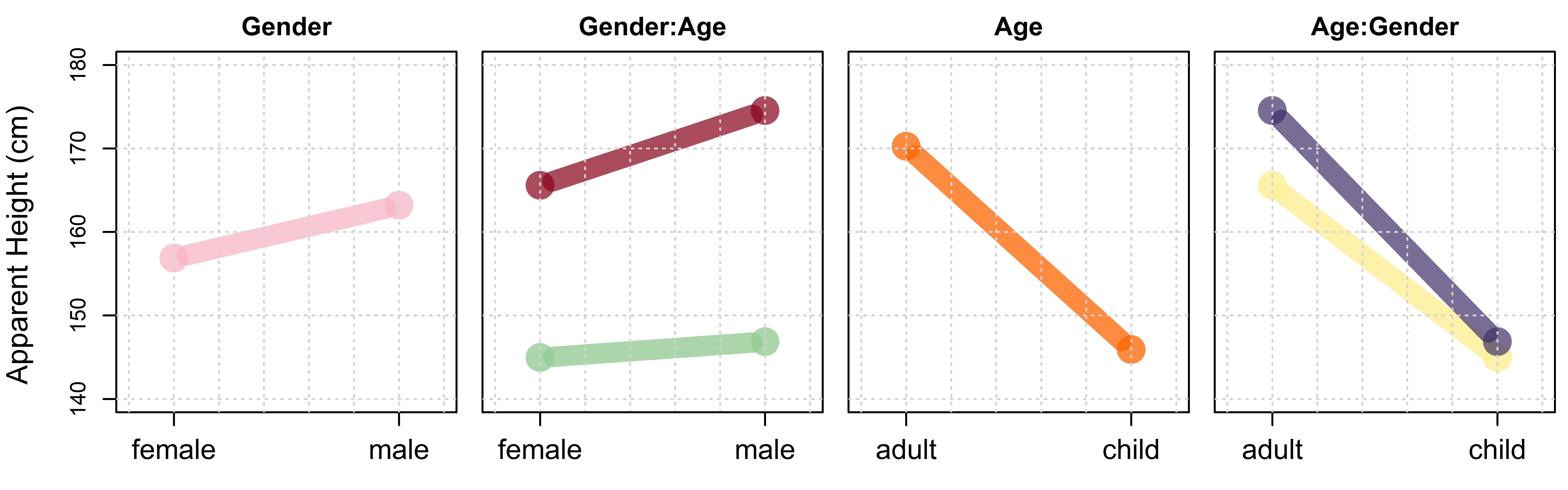
Figure 7.7: Plots showing different ways to consider average apparent height judgments across levels of apparent age and gender.
The plot highlighting the effect of apparent gender on apparent height in figure 7.7 is a main effects plot. Main effects (discussed in chapter 6) are the effects of one predictor averaged across everything else. Saying ‘averaged across everything else’ basically means we are ignoring everything else in our model. A person looking only at the left plot in figure 7.7 would have no idea our data also investigates the effect of apparent adultness. We have ‘erased’ the differences in apparent adultness from our data by averaging across all levels of that factor. Another way to think of main effects is that they are marginal effects, the overall, average, unconditional differences between the levels of the factor. So, if someone asks you “whats the average difference in apparent height between apparent males and females?” you can respond “about 6 cm”. However, as noted in the previous chapter, sometimes the answer is not so simple, and it starts more like: “well… it depends”. Interactions represent situations like these, where the effect of one variable depends on, or is conditional on, the value of some other variable.
The idea of conditional effects (interactions) may feel complicated, but it is something we all understand intuitively. For example, how much will a given person grow in height in the next 5 years? What if I told you that one person is 1 and the other is 60? You know that makes a difference, which is to say, that you know there is an interaction between age and rate of growth. In other words, the expected relationship between time and growth is different for young children and grown adults. Anyone who understands these varying effects understands interactions, whether or not they know how to relate this concept to the mathematical formalisms used to implement these in regression models.
To understand interactions we need to talk about simple effects or simple main effects, the effects of a factor at one specific level of another factor. Basically, main effects show you the overall effect for a factor and simple effects show you the effects of the factor depending on the value of other things. We split the single (main) effect for gender into two simple (main) effects of gender across the levels of apparent age, presented in the second plot in figure 7.7. We can see clearly that the lines do not have the same slope, indicating the effect for gender varies across the levels of apparent age.
The second and fourth plots in figure 7.7 are sometimes called interaction plots. Knowing how to interpret interaction plots is extremely useful because they help you quickly understand the nature of any interactions in your data. Interaction plots show you the simple main effects of one predictor across the levels of the other. They do this by having one factor vary across the x-axis and using different lines for the levels of the other factor. For example in the second plot gender varies across the x axis and different lines are used for the levels of age, and in the fourth plot age varies across the x axis and different lines are used for gender. Note however that the same information is presented in the two interaction plots in figure 7.7, it is just presented differently in each case. In fact, we can see that the y-axis values of the four line endpoints are equal across the two plots. The slopes of the lines in the second plot correspond to the differences between line endpoints in the fourth plot; one is small and one is near zero. The differences between the line endpoints in the second plot are the slopes of the fourth plot, both are large but one is larger.
Imagine what would happen if we began with a main effect plot like the first plot in figure 7.7, and added/subtracted a single value to each end of the line segment. This single value could represent the main effect of some other factor, for example the main effect for apparent age. Since a single number would be added to both points on the line, the line representing the effect of apparent gender would slide up/down the y-axis, however, the line would not change its slope. We might imagine trying to slide the line in the first plot in figure 7.7 up and down the y axis to try to get it to match the simple effects presented in the second plot of the same figure. However, we will not be successful because these two lines do not share a slope. In order to make the lower and upper lines not be parallel, we need to add different values to each side of the main effects line.
The varying effect for apparent gender at different levels of apparent age indicates that these effects interact in our data. Interactions between our factors are detectable visually in the form of a lack of parallel lines in interaction plots (i.e. varying slopes in the simple effects). In the absence of an interaction, all lines in such a plot should be parallel, save for variation due to random error. We can see that the lines in the second and fourth plots in figure 7.7 are not parallel, suggesting an interaction between apparent age and gender in our data. In the absence of an interaction, we could just answer the question “whats the average difference between apparent height between males and females?” with a number like 6 cm. In the presence of an interaction we need to consider the conditional effects of each predictor at the levels of the other predictor (i.e., the simple effects). So, the most important thing to remember about interpreting main effects is:
If there are no interactions between factors, focus on main effects.
If there are interactions between factors, you must inspect the simple effects to understand the nature of the interaction.
7.6 Investigating interactions with a model
In figure 7.6 we made posterior predictions of our data with and without the inclusion of the speaker random effects, and presented these as boxplots. We present these same predictions as interaction plots in figure 7.8. We can see in figure 7.8 that the predictions made using only fixed effects (middle plot) are systematically wrong, and that the problem with our predictions is that our predicted lines are parallel, while those for our real data are not. As we’ve just discussed, in the absence of interactions, interaction plots contain only parallel lines. Since our model (model_both) does not include interaction terms it cannot represent interactions, and so is only capable of making predictions along parallel lines. This means our model is not capable of representing the pattern in our data using only fixed effects.
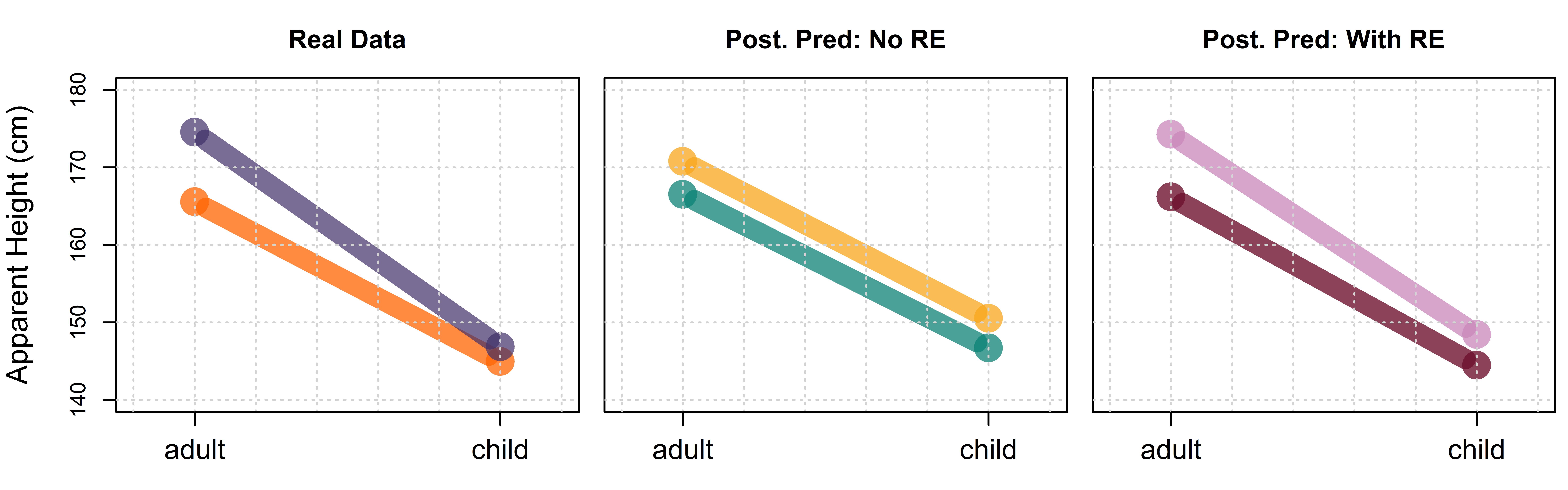
Figure 7.8: Interaction plots comparing our real data to posterior predictions, with and without random effects (RE). In each case the top line represents height judgments for apparent males.
We can see that when random effects are included, prediction is better. This is not surprising since the listener and speaker-specific intercept adjustments allow for deviations from the fixed effects predictions to still be captured by the model. However, if accurate prediction is only possible using random effects, that limits the generalizability of our model to new levels of the grouping variable, e.g. subject or listener. In other words, the absence of good fixed-effects prediction limits the generalizability of our model and may be a cause for concern.
Our model is a little universe we made up, and it only includes the information, and structures, we included in it. This ‘universe’ only contains parallel lines because we only gave it that capability. So, the fact that our model generates parallel lines does not in any way ‘prove’ that the lines are parallel, because they were bound to be. This is a little bit like taking black and white photographs and then using this as evidence that the scene does not contain color. In order to properly investigate whether the lines are parallel, the model must be built in a way that allows it to (at least potentially) represent the interactions in our data, i.e., to represent lines that are not parallel.
Before fitting a model with an interaction term we want to remind you that you cannot estimate interactions between factors that are not crossed: If you did not observe all combinations of factor levels, you are not in a position to talk about how effects vary for those combinations. So, it is extremely important to be sure to only include interactions in your model when these predictors are crossed, or your model may run into all sorts of problems.
7.6.1 Data and research questions
We load our data and packages again (again) just in case:
library (brms)
library (bmmb)
options (contrasts = c('contr.sum','contr.sum'))
data (exp_data)We’re going to a use the very same variables from our data frame that we used in the previous model. These are:
L: A number from 1-15 indicating which listener responded to the trial.height: A number representing the height (in centimeters) reported for the speaker on each trial.S: A number from 1-139 indicating which speaker produced the trial stimulus.G: The apparent gender of the speaker indicated by the listener,f(female) orm(male).A: The apparent age of the speaker indicated by the listener,a(adult) orc(child).
And we’re going to address our two previous research questions, and add one more:
(Q1) Does average apparent height differ across levels of apparent age?
(Q2) Does average apparent height differ across levels of apparent gender?
(Q3) Is there an interaction between the effects of apparent gender and age on apparent height?
7.6.2 Model formulas
The model presented above (model_both) requires only a slight tweak to include the interaction in our data. There are two ways to include interactions in R model formulas, either:
height ~ A + G + A:G + (A + G + A:G|L) + (1|S)
Or:
height ~ A * G + (A * G|L) + (1|S)
The first way includes an explicit interaction term, A:G. The syntax for these is X:Z for an interaction between effects X and Z, W:X:Z for a three-way interaction, and so on. The second way uses * between our two predictors. This tells R to include those predictors, and all the interactions between them. This can be much faster than specifying all interactions, but you lose control over which ones you include. For example, this formula:
y ~ Z * X * W
Implies this one:
y ~ Z + X + W + Z:X + Z:W + X:W + Z:X:W
But cannot represent this one (since it omits the Z:W interaction):
y ~ Z + X + W + Z:X + X:W + Z:X:W
Actually, we could represent the model above using the following formula:
y ~ Z * X * W - Z:W
Where we use - to remove specific terms from the model formula. We can also use parentheses in model formulas to organize our interactions as in the following formula:
y ~ (Z + X) * W
Which is equivalent to:
y ~ Z + X + W + Z:W + X:W
We can also use ^ to find all interactions up to a certain order/degree, as in:
y ~ (Z + X + W)^2
y ~ (Z + X + W)^3
Which are equivalent to:
y ~ Z + X + W + Z:X + Z:W + X:W
y ~ Z + X + W + Z:X + Z:W + X:W + Z:X:W
Finally, all of this applies to both our fixed and random effects predictors as in the following possible formula:
y ~ (Z + X) * W * V - W:V:X + ((Z + X) * W * V - W:V:X|L)
In our formulas, the : and | both basically mean given, and indicate that conditional effects are being estimated. The difference between these is syntactic, much like the difference in R between [] and (). A colon (:) is used to indicate interactions between ‘fixed’ effects, that is effects estimated using fixed prior distributions. The pipe (|) indicates that the formula to its left is being estimated for each level of the predictor to the right of the pipe, and that this estimation is being carried out using adaptive partial pooling. In the formula above, the use of : and | for different purposes makes ((Z + X) * W * V - W:V:X|L) interpretable: It means that, effectively, sub-models equivalent to (Z + X) * W * V - W:V:X will be fit for each level of L.
7.6.3 Description of our model
Our full model specification is presented in (7.8), once again omitting the equation specifying \(\mathrm{\Sigma}\). The model now includes an interaction term (\(A \colon G\)) that can help explain variation that cannot be explained by the independent effects of adultness and gender. This interaction term helps us model the conditional effect of one predictor given the other. Our model also actually contains a three-way interaction as well (\(A \colon G \colon L_{[\mathsf{L}_{[i]}]}\)), though it may not seem that way to us superficially. This is the ‘random’ effect of the interaction between \(A\) and \(G\) for each level of listener, (A:G|L) in our model. This is a three-way interaction because it represents the interaction between the two-way interaction of \(A\) and \(G\) with our listener factor: The listener dependent two-way interaction between \(A\) and \(G\).
\[ \begin{equation} \begin{split} height_{[i]} \sim \mathrm{t} (\nu, \mu_{[i]},\sigma) \\ \mu_{[i]} = \mathrm{Intercept} + A + G + A \colon G + \\ L_{[\mathsf{L}_{[i]}]} + A \colon L_{[\mathsf{L}_{[i]}]} + G \colon L_{[\mathsf{L}_{[i]}]} + A \colon G \colon L_{[\mathsf{L}_{[i]}]} + S_{[\mathsf{S}_{[i]}]} \\ \\ \mathrm{Priors:} \\ S_{[\bullet]} \sim \mathrm{t}(3,0,\sigma_S) \\ \begin{bmatrix} L_{[\bullet]} \\ A \colon L_{[\bullet]} \\ G \colon L_{[\bullet]} \\ A \colon G \colon L_{[\bullet]} \end{bmatrix} \sim \mathrm{MVNormal} \left( \begin{bmatrix} 0 \\ 0 \\ 0 \\ 0 \\ \end{bmatrix}, \mathrm{\Sigma} \right) \\ \\ \\ \mathrm{Intercept} \sim \mathrm{t}(3,156,12) \\ A, G, A \colon G \sim \mathrm{t}(3,0,12) \\ \sigma_L, \sigma_{A \colon L}, \sigma_{G \colon L}, \sigma_{A \colon G \colon L}, \sigma_S \sim \mathrm{t}(3,0,12) \\ \sigma \sim \mathrm{t}(3,0,12) \\ \nu \sim \mathrm{gamma}(2, 0.1) \\ R \sim \mathrm{LKJCorr} (2) \end{split} \tag{7.8} \end{equation} \]
Our updated plain English description now says:
We are modeling apparent height as coming from a t distribution with unknown nu (\(\nu\)), mean (\(\mu\)), and scale (\(\sigma\)) parameters. The expected value for any given trial (\(\mu\)) is modeled as the sum of an intercept, an effect for apparent speaker age (\(A\)), gender (\(G\)), and their interaction (\(A \colon G\)), a listener effect (\(L\)), a listener dependent effect for apparent age (\(A \colon L\)), apparent gender (\(G \colon L\)), their interaction (\(A \colon G \colon L\)), and a speaker effect (\(S\)). The speaker effects were drawn from a univariate normal distribution with a standard deviation (\(\sigma_S\)) estimated from the data. The four listener effects were drawn from a multivariate normal distribution with individual standard deviations (\(\sigma_L,\sigma_{A \colon L},\sigma_{G \colon L},\sigma_{A \colon G \colon L}\)) and a correlation matrix (\(R\)) that was estimated from the data. The remainder of the ‘fixed’ effects and correlations were given prior distributions appropriate for their expected range of values.
Before continuing, we want to address the fact that we’ve included the same structure in our ‘fixed effects’ as for our listener ‘random effects’. We will do this for all the models we fit in this book whenever possible. There are a couple of reasons for this, and we will discuss these with respect to the ‘random effect’ of apparent age by listener (i.e. the \(A \colon L\) interaction). Not including this predictor is equivalent to setting it to zero for all listeners (and setting \(\sigma_{A \colon L}=0\)), meaning we expect the effect for apparent age to be the same across all levels of listener. However, we saw in the previous chapter that this is not the case and that there is substantial variation in the effect for apparent age across listeners. So, omitting random effects relies on the assumption that the variation across listeners (or whatever variable defines the random effects) is equal to zero, which may not be well founded a priori.
Furthermore, in chapter 6 we fit models with and without random effects and saw that the model with random effects produced substantially wider credible intervals for the apparent age ‘fixed’ effect, reflecting increased uncertainty in the parameter estimate. In that case, assuming listener-variation in the parameter was zero when it was not resulted in an overly precise estimate of the apparent age parameter. So, we see that omitting random effects means assuming they are equal to zero, and can affect the amount of uncertainty we get in our resulting parameter estimates. The only way to know whether a listener-dependent parameter shows substantial variation or not is to include the possibility for listener-dependent variation in your model, at least initially. Of course, the inclusion of any ‘random effect’ in your model assumes that: 1) Your model is designed in a way that the ‘random’ effect can be estimated, and 2) You have enough data to estimate those parameters. These issues will be discussed further in chapter 11.
7.6.4 Fitting and interpreting the model
Below we fit the model including an interaction term. Remember that the line set_prior("student_t(3, 0, 12)", class = "b") sets the prior for all non-intercept ‘Population-Level’ predictors (i.e. fixed effects). This includes all of our main effects predictors and our interaction terms.
# Fit the model yourself
priors = c(brms::set_prior("student_t(3,156, 12)", class = "Intercept"),
brms::set_prior("student_t(3,0, 12)", class = "b"),
brms::set_prior("student_t(3,0, 12)", class = "sd"),
brms::set_prior("lkj_corr_cholesky (2)", class = "cor"),
brms::set_prior("gamma(2, 0.1)", class = "nu"),
brms::set_prior("student_t(3,0, 12)", class = "sigma"))
model_interaction =
brms::brm (height ~ A + G + A:G + (A + G + A:G|L) + (1|S),
data = exp_data, chains = 4, cores = 4, warmup = 1000,
iter = 5000, thin = 4, prior = priors, family = "student")# Or download it from the GitHub page:
model_interaction = bmmb::get_model ('7_model_interaction.RDS')We are primarily interested in discussing interactions, but we want to spend a moment looking at the listener random effects in our model, since there are now several.
# inspect fixed effects
bmmb::short_summary (model_interaction)
## Formula: height ~ A + G + A:G + (A + G + A:G | L) + (1 | S)
##
## Group-Level Effects:
## ~L (Number of levels: 15)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 4.24 0.79 2.96 6.03
## sd(A1) 4.47 0.85 3.13 6.42
## sd(G1) 2.10 0.49 1.37 3.22
## sd(A1:G1) 1.34 0.36 0.76 2.13
## cor(Intercept,A1) -0.71 0.13 -0.90 -0.38
## cor(Intercept,G1) -0.20 0.22 -0.59 0.24
## cor(A1,G1) -0.24 0.21 -0.62 0.20
## cor(Intercept,A1:G1) 0.17 0.23 -0.29 0.58
## cor(A1,A1:G1) -0.02 0.23 -0.46 0.43
## cor(G1,A1:G1) -0.34 0.24 -0.74 0.18
##
## ~S (Number of levels: 139)
## Estimate Est.Error l-95% CI u-95% CI
## sd(Intercept) 2.36 0.31 1.79 2.99
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI
## Intercept 158.46 1.12 156.22 160.62
## A1 10.78 1.21 8.39 13.15
## G1 -2.87 0.60 -4.07 -1.72
## A1:G1 -1.64 0.41 -2.44 -0.81
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI
## sigma 5.01 0.16 4.70 5.34
## nu 3.44 0.33 2.87 4.15We can see in the short model summary above that we’ve estimated four standard deviation terms for our listener-dependent predictors. These correspond to \(\sigma_{L}\) (sd(Intercept)), \(\sigma_{A \colon L}\) (sd(A1)), \(\sigma_{G \colon L}\) (sd(G1)), and \(\sigma_{A \colon G \colon L}\) (sd(A1:G1)). We’ve also estimated six correlation terms for our listener random effects. Recall that our random effects are drawn from a multivariate normal distribution. We have four terms which means this four-dimensional distribution is associated with a 4$$4 correlation matrix and 16 correlation terms. Of the correlations, four (on the main diagonal) equal one and can be ignored. This leaves 12 terms. Of these, only half are unique because the correlation of \(x\) and \(y\) equals the correlation of \(y\) and \(x\). This is how we end up with six estimated correlations for our four listener-dependent predictors.
We can now focus on the fixed effects (Population-Level Effects), of which there are four: An intercept, two main effects terms and an interaction. The interaction term is just another element of your prediction equation intended to help explain variation that can’t be predicted by the main effects of predictors involved in the interaction. If there is no interaction in your data, then the value of the interaction term will be close to zero and your model will look just as if you had not included the interaction at all. A look at the model output above indicates that our interaction is much smaller than the age effect, but not much smaller than the gender effect. This suggests it has the potential to influence our conclusions about gender more than our conclusions about age.
Why is there only a single interaction? Basically for the same reason that we can only estimate a single effect for apparent age and gender (i.e., linear dependence, discussed in section 5.6 and 8.8). The number of levels of a factor that you can estimate is generally one fewer than the number of levels in the factor. For interaction terms, the number of parameters you can estimate is equal to (number of levels of factor A - 1)\(\times\)(number of levels of factor B - 1) for two factors. Since each of our factors have two levels, we can only estimate one parameter, \((2-1) \times (2-1)=1\).
The A1:G1 term can be read aloud as “the effect of A1 given (conditional on) the level of G1”. For individual factors, our models cannot estimate A2 because -A1 = A2. For interactions between factors with two levels, the model cannot estimate A2:G1 because -A1:G1 = A2:G1. For interactions between factors with two levels, every time you want to change A1 or G1 to A2 or G2 you flip the sign of the A1:G1 term instead. As a result, going left to right and changing one level at a time we get A1:G1 = -A2:G1 = A2:G2 = -A1:G2, which is precisely why we can’t estimate more than one interaction for this model. Why does this happen? Looking at how interaction plots are built up out of main effects and interactions may help illustrate the reason for this.
Figure 7.9 presents an example of how our interaction plot can be built up from the components in our fixed effects above, and is very useful for understanding the geometry of main effects and interactions. In the top left plot we see that the intercept lifts up a line from 0 to the level of the overall mean apparent height of 158 cm. Then, the effect for A1 (adult, +11 cm) and -A1 (children, -11 cm) are added to this value, causing the separation between the lines indicating a ‘main effect’ for adultness. Notice that the lines are parallel with respect to each other, and also parallel to the x-axis. This is because we have not added an effect for gender, which will be indicated as a slope along the x axis. In the middle plot in the top row we see the addition of the gender effect: G1 (female, -2.8 cm), and -G1 (male, 2.8 cm). Notice that the effect is added to one end of each line segment and subtracted from the other end of the line segment. This is what causes a (single) slope along the x axis for both lines. The right plot in the top row shows the addition of all of the effects shown in the top row, the result of a model that includes both main effects but the same non-zero slope for both lines (i.e. no interaction yet).
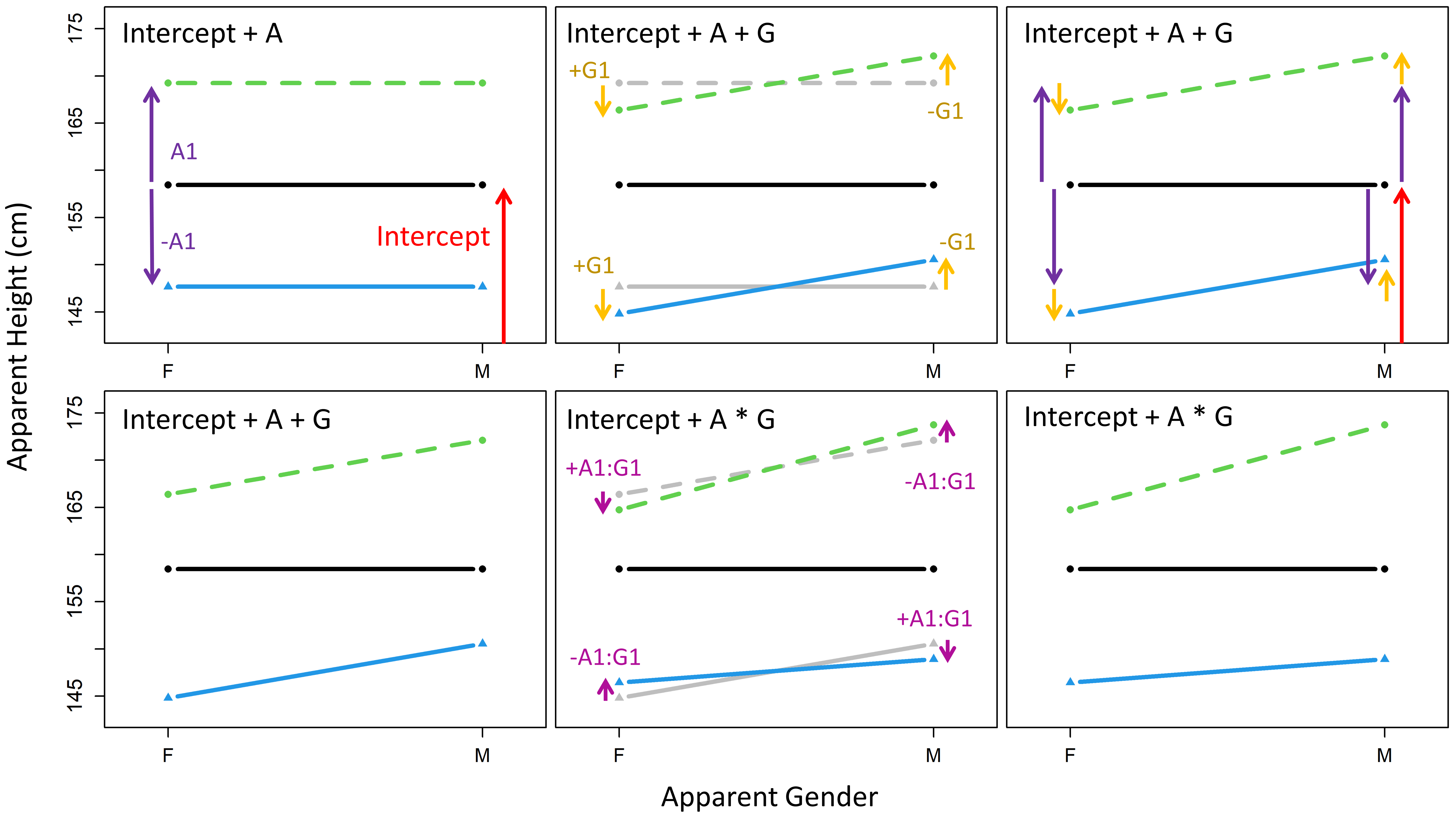
Figure 7.9: An interaction plot built up from its individual components.
In the middle plot in the bottom row of figure 7.9, we add the gender interaction A1:G1, which was -1.6 cm, to each end point for both lines. Notice that the interaction effect has different signs when added to the same line, and also for each end of each line segment. In each case, the sign is determined by the specific speaker category it is applied to. For example, we know that A1:G1 represents female adults (top left point). So, we flip the sign for adult children in the bottom-left point (i.e. -A1:G1 = A2:G1), and we flip the sign again for male children in the bottom right point (i.e. A1:G1 = A2:G2). The sign flips result in opposing interactions at each x axis position across lines, but also for different endpoints along the same line. We can see that beginning with the parallel lines in bottom-left, the interaction added in the bottom middle plot constitutes an equal ‘pinch’ of one end of the lines and a spreading of the other side of the lines. This equal pinch and spreading is necessary in order to maintain the same average slope across both lines (the gender main effect) and the same average spacing between both ends of the lines (the age main effect). This is effectively why we get only a single interaction term in a model with this fixed effects structure.
7.6.5 Caulculating group means in the presence of interactions
Recovering the predicted group means using the model coefficients is straightforward, but can be tedious for larger models: We must now either add or subtract the value of the interaction term (A1:G1) from each group as determined by the combination of main effects. We can easily determine which to do for this model because the sign on the interaction term is the product of the signs on the relevant ‘main effects’ terms. So, two positive or negative main effects result in a positive interaction term, while mismatched positive and negative main effects result in a negative interaction.
Below, we use short_hypothesis to reconstruct expected group means given the model fixed effects. The fourth hypothesis we’re testing below is the simplest to understand, so we will start there. This hypothesis asks whether the sum of the Intercept, A1 (the effect for adultness), G1 (the effect for femaleness), and A1:G1 (the interaction of adult and female), is equal to zero. Because of the specific parameters we combined, this results in the comparison of the mean height for women to zero. The first hypothesis below says Intercept + -A1 + -G1 + A1:G1 = 0. Since we flipped the sign on A1 and G1, these now correspond to the effects of ‘child’ and ‘male’ (i.e. boy). Since both A1 and G1 are negative, we flip the sign on the interaction term twice and it remains positive. Note that for all of our hypotheses below, the sign on the interaction terms always depends on the signs of the corresponding main effects terms.
# intercept, boys, girls, men, women
means_pred_interaction = bmmb::short_hypothesis (
model_interaction,
c("Intercept + -A1 + -G1 + A1:G1 = 0", # boys
"Intercept + -A1 + G1 + -A1:G1 = 0", # girl
"Intercept + A1 + -G1 + -A1:G1 = 0", # men
"Intercept + A1 + G1 + A1:G1 = 0")) # womenBelow we compare the actual mean apparent height for each group to the predictions made without and with an interaction term. We can see that in each case, the prediction made by the model with an interaction are closer to the data means than those made by the model without an interaction. In addition, the predictions made including an interaction finally do reflect the fact that the difference in apparent height between apparent boys and girls is much smaller than that between apparent men and women.
# actual data means
tapply (exp_data$height, exp_data$C, mean)
## b g m w
## 146.9 145.0 174.5 165.6
# predictions with no interaction term
means_pred[,1]
## [1] 150.9 146.5 170.8 166.4
# predictions with interaction term
means_pred_interaction[,1]
## [1] 148.9 146.4 173.8 164.77.6.6 Calculating simple effects in the presence of interactions
We know that we have an interaction in our data, and that this interaction needs to be investigated by considering the simple effects for each factor. Below we use short_hypothesis to find the simple effects of age across levels of gender, and of gender across levels of age. To recover the simple effect for a predictor, we add the main effect term to the appropriate interaction term (including the appropriate sign). To figure out which sign to use, you just need to think about which group is referred to by each interaction. For example, in the first line below we add A1, the age main effect, to A1:G1. Since it has a G1 in it and we are adding and not subtracting, we know this refers to female speakers. As a result, A1 + A1:G1 results in the simple effect for apparent age for apparent females. In the second line we again use A1 but now add -A1:G1, which we know is equal to A1:G2 (which does not exist). As a result, A1 - A1:G1 results in the simple effect of apparent age for apparent males.
# intercept, boys, girls, men, women
simple_effects = bmmb::short_hypothesis (
model_interaction,
c("A1 + A1:G1 = 0", # effect for apparent age for adults (G1)
"A1 - A1:G1 = 0", # effect for apparent age for children (-G1)
"G1 + A1:G1 = 0", # effect for apparent gender for adults (A1)
"G1 - A1:G1 = 0")) # effect for apparent gender for children (-A1)
# predictions with interaction term
simple_effects
## Estimate Est.Error Q2.5 Q97.5 hypothesis
## H1 9.142 1.2792 6.639 11.5977 (A1+A1:G1) = 0
## H2 12.424 1.2707 9.938 14.9913 (A1-A1:G1) = 0
## H3 -4.514 0.6174 -5.731 -3.3177 (G1+A1:G1) = 0
## H4 -1.232 0.8192 -2.860 0.3919 (G1-A1:G1) = 0We will provide a verbal description of the calculations involved in estimating simple effects. We will use our fixed effects estimates, rounded to make the discussion easier to follow. We’ve previously warned against averaging parameters first and combining them second. However, this approach can be useful for a quick investigation of our model parameters and is fine for our purposes.
round (brms::fixef (model_interaction))
## Estimate Est.Error Q2.5 Q97.5
## Intercept 158 1 156 161
## A1 11 1 8 13
## G1 -3 1 -4 -2
## A1:G1 -2 0 -2 -1First, we will calculate the simple effects for apparent age at the levels of apparent gender. The average apparent height across all groups is 158 cm. There is an age-based 11 cm deviation from the intercept, meaning the difference between apparent adults and children is 22 cm (11 \(\cdot\) 2). This means that the averages for apparent adults and children are about 169 and 147 cm (158 ± 11). However, the A1:G1 interaction is -2 cm. This means that when the speaker was identified as female (G1), the affect for adultness decreased from 11 cm to 9 cm (11 - 2, A1+A1:G1), meaning the difference across women and girls was 18 cm (2 \(\cdot\) 9). In contrast, when the speaker was identified as male (-G1), the affect for adultness increased from 11 cm to 13 cm (11+2, A1-A1:G1), meaning the difference between boys and men was about 26 cm. Notice that in the calculations above, the effect for apparent age would be equal across apparent genders in the case that there is no interaction (i.e. A1:G1=0).
We could instead consider the simple effects for apparent gender given apparent age. There is a gender-based 3 cm deviation from the intercept, meaning the difference between apparent adults and children is -6 cm (-3 \(\cdot\) 2). This means that, overall, the female and male averages are about 155 and 161 cm respectively (158 ± -3). However, the A1:G1 interaction is -2 cm. This means that when the speaker was an adult (A1) the effect for gender increased in magnitude from -3 cm to -5 cm (-3 + -2, G1+A1:G1), resulting in a difference between groups of 10 cm (-5 \(\cdot\) 2). However, when the speaker was a child (-A1), the effect for gender decreased in magnitude from -3 cm to 1 cm (-3+2, G1-A1:G1), resulting in a difference in groups of 2 cm based on apparent gender (1 \(\cdot\) 2).
7.6.7 Assessing model fit: Bayesian \(R^2\)
We can assess the fit for our model including interaction terms by making more posterior predictions with our new model. We will focus on the average predictions for each data point since our objective is to see to what extent the fixed effects can represent the patterns in the data.
y_post_pred_int = predict (model_interaction)
y_post_pred_no_re_int = predict (model_interaction, re_formula = NA)In figure 7.10 we compare our data, the predictions of our original model, and the predictions of our model that includes interactions (both models with and without random effects). Whereas the model with no interactions enforced parallelism on the simple effects, our new model is able to capture the interaction in our data by representing different effects for gender at different levels of age (and vice versa). Although both of our models are able to capture the pattern in the data when random effects are included, only one of these is able to do so with its fixed effect structure.
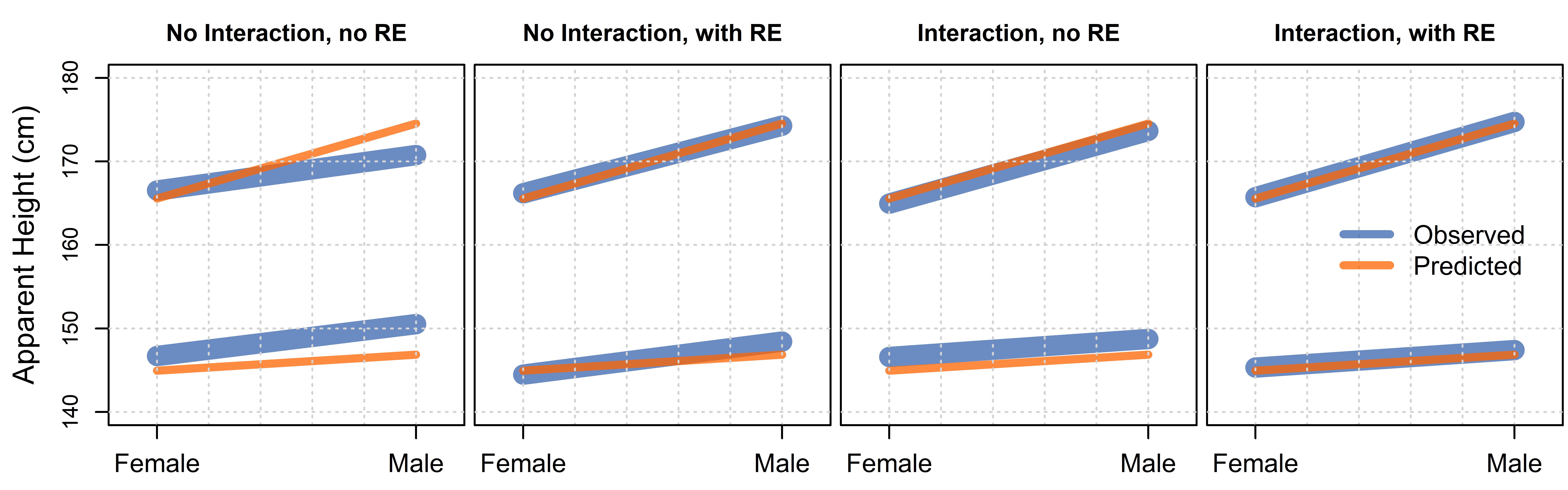
Figure 7.10: Interaction plots comparing observed apparent height judgmentss to different posterior predictions. Posterior predictions with no interaction come from model_both and predictions with interactions come from model_interaction. With RE (random effects) refers to predictions made using random effects, and no RE refers to predictions made without random effects.
We can compare the models with and without the interaction term using leave one out cross-validation, discussed in section 6.4.3.
model_both = brms::add_criterion (model_both, criterion="loo")
model_interaction = brms::add_criterion (model_interaction, criterion="loo")The comparison suggests a large difference between the models, but also some uncertainty regarding the difference.
brms::loo_compare (model_both, model_interaction)
## elpd_diff se_diff
## model_interaction 0.0 0.0
## model_both -29.1 11.2To some extent the difference may appear to be smaller than it is because our random effects do a good job of explaining listener and speaker-dependent deviations from the expectations set out by the fixed effects. Explaining things with ‘random’ effects is less useful for understanding the out-of-sample characteristics of your data. If we were to replicate this experiment with new speakers and listeners, we would expect all the random effects to change. However, if the relations between apparent age and gender and apparent height expressed in our model are ‘true’, we would expect the ‘fixed’ effects in the replication to be about the same. For this reason, it is extremely useful to consider fixed effects prediction in addition to the predictive power of a complete model including both fixed and random effects.
To quantify how well our models fit our data we can also use Bayesian \(R^2\). There are potentially several ways to define \(R^2\) in a Bayesian context. The account of Bayesian \(R^2\) presented here is a summary of the proposal outlined in Gelman et al. (2019). In addition, the discussion of \(R^2\) to follow assumes that your model involves either Gaussian or t-distributed errors and a continuous dependent variable.
\(R^2\) is a measure of model fit that breaks down the total variance in your dependent variable into two parts: 1) The variance that your model can explain, and 2) the variance that your model cannot explain (the residual error, which we’ve been calling \(\sigma\)). This is shown in (7.9).
\[ \begin{equation} \sigma^2_{total} = \sigma^2_{explained} + \sigma^2_{error} \tag{7.9} \end{equation} \]
\(R^2\) is the ratio of the explained variance to the total variance, as seen in (7.10). Since the explained variance cannot be less than zero nor can it be greater than the total variance, values of \(R^2\) range from zero to one. As a result, \(R^2\) can be interpreted as the proportion of variance in the dependent variable that is explained by the model
\[ \begin{equation} R^2 = \frac{ \sigma^2_{explained} }{ \sigma^2_{total} } = \frac{ \sigma^2_{explained} }{ \sigma^2_{explained} + \sigma^2_{error} } \tag{7.10} \end{equation} \]
To make \(R^2\) a Bayesian \(R^2\), we need to incorporate the posterior distribution of parameter values into our estimation. This is done, in part, by calculating a value of \(R^2\) for every individual set of posterior samples, as shown in (7.11). For every posterior sample of parameter values \(s\) (from \(S\) total samples) we calculate: 1) The variance in the predicted values (\(\hat{y}\)) of the dependent variable for our \(n\) data points (\(V_{i=1}^n \; \hat{y}^s_n\)), and 2) the variance of the residual error for our \(n\) data points (\(V_{i=1}^n \; \hat{e}^s_i\)).
\[ \begin{equation} R^2_s = \frac{V_{i=1}^n \; \hat{y}^s_i}{ V_{i=1}^n \; \hat{y}^s_i + V_{i=1}^n \; \hat{e}^s_i} \tag{7.11} \end{equation} \]
The residual error is the difference between predicted (\(\hat{y}\)) and observed (\(y\)) values for each data point, shown in (7.11). Notice that the predictions and residuals vary across the \(s\) samples, but our observed data does not.
\[ \begin{equation} \hat{e}^s_i = \hat{y}^s_i - y_i \tag{7.12} \end{equation} \]
This results in a distribution of \(R^2\) values, unlike more ‘traditional’ estimates of \(R^2\) that result in point estimates. The brms package includes a function called bayes_R2 that helps you calculate Bayesian \(R^2\) for brm models. However, we’re going to use the r2_bayes function from the bmmb package instead because this function allows you to modify (or omit) the fixed effects included in the prediction. Below we calculate \(R^2\) for the models we fit this chapter, including all random effects.
r2_both = r2_bayes(model_both)
r2_interaction = r2_bayes(model_interaction)We can inspect the output and see that they have very similar values. This is not surprising given their similarities seen in figure 7.10.
r2_both
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.7755 0.005566 0.7641 0.7858
r2_interaction
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.7802 0.005347 0.7694 0.7901Just as we can omit the random effects when we use predict, we can omit random effects from our calculations of \(R^2\) by setting re_formula = NA.
r2_both_no_re = r2_bayes(model_both, re_formula = NA)
r2_interaction_no_re = r2_bayes(model_interaction, re_formula = NA)Now a comparison reveals a more substantial difference in explanatory power between the models.
r2_both_no_re
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.503 0.07258 0.3411 0.6216
r2_interaction_no_re
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.5732 0.05909 0.4389 0.6678We would rather explain as much variation as possible using fixed as opposed to random effects. In addition, the interactions ‘makes sense’ given our domain knowledge. What we mean by this is that we know that the difference in veridical height between boys and girls 10-12 years old is inconsistent and may be about zero on average. So, we should not be too surprised if the effect for apparent gender on apparent height is small for apparent children. However, there is a larger and more consistent difference in average veridical height between adult males and females. As a result, we do expect a meaningful effect for apparent gender on the apparent heights of adults. For these reasons, it would be surprising if there were not an interaction, and we prefer the model with the interaction despite the middling support provided by our cross-validation model comparison.
\(R^2\) is not really going to prove anything, and shouldn’t form the basis of your model selection. We believe that it is a useful method to be aware of in addition to, and not instead of, other approaches such as \(\mathrm{elpd}\)). The primary utility of \(R^2\) is that since it reflects a proportion between 0 and 1, it is more easily interpretable for humans. For example, the difference in \(\mathrm{elpd}\) between model_both and model_interaction is about 29, which is large enough to be statistically meaningful. However, how much does this difference actually matter at the level of our dependent variable? Well, the proportion of variance explained by the two models is only different by about 0.5%, suggesting that these models do not offer substantially different predictions. In contrast, we might imagine the same difference in \(\mathrm{elpd}\) being associated with a 10% change in the variance explained, which will likely be a noticeable difference. In this way, \(R^2\) can be a useful indicator of how much our predictions are really changing, provided in a measure that many readers will find relatively intuitive.
7.7 Answering our research questions
We will answer our research questions based on model_interaction. In addition, we used the forpaper function, like this forpaper(fixef(model_interaction)) to generate standard output of the form (mean = --, s.d. = --, 95% C.I = [--, --]) from the typical brms coefficient table output.
Results indicate that the average apparent height across all speaker groups (i.e. the intercept) was 158.5 (s.d. = 1.12, 95% C.I = [156.22, 160.62]). We also found an average effect of 10.8 cm for apparent speaker age (s.d. = 1.21, 95% C.I = [8.39, 13.15]) and -2.9 cm for apparent speaker gender (s.d. = 0.6, 95% C.I = [-4.07, -1.72]). In addition, we found an interaction between the effects of apparent age and apparent gender on apparent heights (mean = -1.64, s.d. = 0.41, 95% C.I = [-2.44, -0.81]). The result of these effects are that apparent adults were perceived as taller than apparent children and apparent males were perceived as taller than apparent females. However, the difference in apparent height due to apparent gender was larger for adults than for children (and the effect for apparent age was larger for males than for females). Figure 7.11 presents the model fixed effects other than the intercept (whose value is too large to plot in this range).
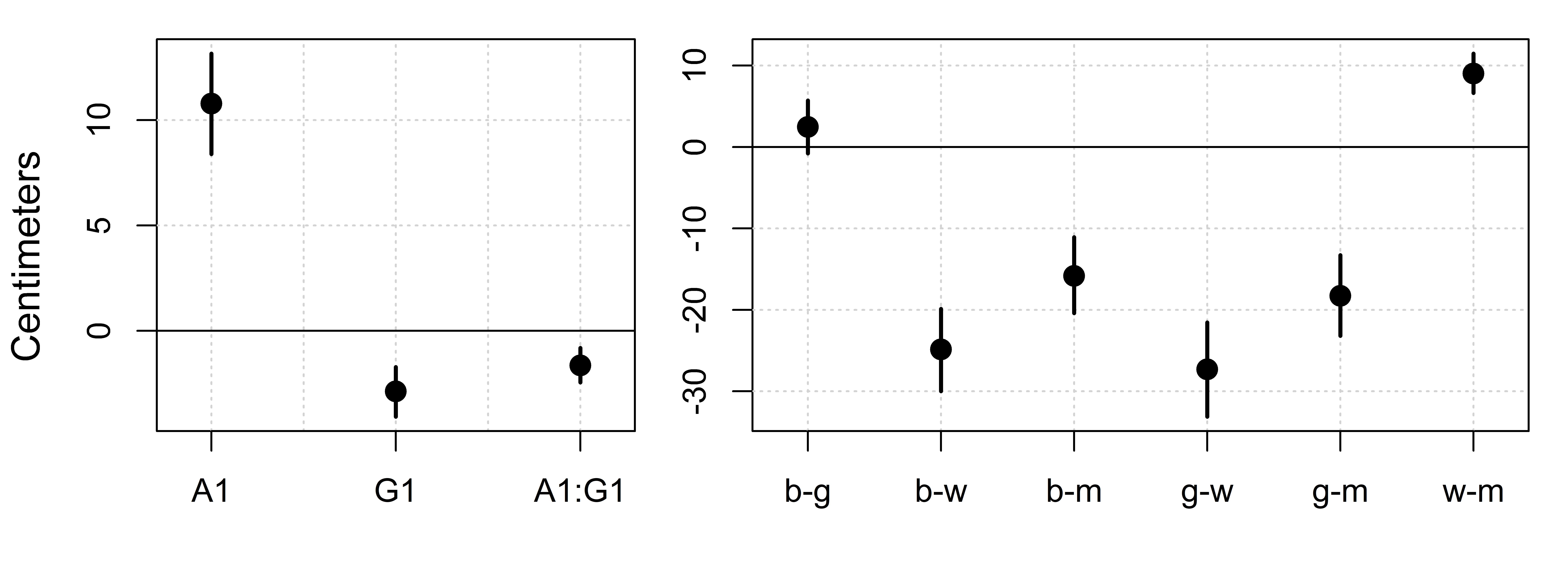
Figure 7.11: (left) Fixed effect means and 95% credible intervals (minus the intercept) for model_interaction. (right) Means and 95% credible intervals for the pairwise differences between predicted means for different apparent speaker categories (boys, girls, men and women).
The fixed effects all have 95% credible intervals that do not overlap with zero. Historically, this sort of result has been thought of as indicating that the effect is ‘real’, as in not equal to zero. In contrast, when the 95% (or some other) credible interval of an effect overlaps with zero, researchers will often conclude that this effect is not ‘real’, i.e. that it is exactly equal to zero. Gelman et al. (2012) note that effects are rarely exactly equal to zero, and that an inability to conclusively distinguish the value of an effect from zero is not the same thing as thinking that the most probable value of an effect is exactly zero.
As a result, rather than focusing on whether any given interval crosses zero or not, Gelman and Carlin (2014) recommend thinking about data analysis in terms of avoiding type S and type M errors. Type S (sign) errors occur when you think an effect is negative but its actually positive (or vice versa). A type M (magnitude) error is when you think an effect is large but it’s actually small (or vice versa). Focusing on type M and S errors rather than whether an interval crosses zero forces researchers to focus on the magnitude of the predictor, its practical importance, and the uncertainty involved in the estimate. That being said, if a parameter has a ‘large’ magnitude (in the context of the dependent variable) and a ‘small’ amount of uncertainty (relative to the magnitude), this predictor is likely to be important for understanding the dependent variable in the context of your model.
Figure 7.11 also presents the pairwise difference between all four apparent speaker groups (boys, girls, women, and men). The difference between pairs of group means is calculated by extracting the samples from the group means we reconstructed in section 7.6.5 (means_pred_interaction). The result of this is a matrix with four columns, each representing a group effect, with individual posterior samples differing along rows. To find the difference between group means between each pair of groups, we subtract the columns representing the group effects and inspect the distribution of the differences. This process is shown below and the distribution of pairwise group differences is presented in the right plot of figure 7.11.
# get group mean predictions
C = attributes(means_pred_interaction)$samples
# find pairwise differences
pairwise_diffs = cbind("b-g"=C[,1]-C[,2], "b-w"=C[,1]-C[,3],
"b-m"=C[,1]-C[,4], "g-w"=C[,2]-C[,3],
"g-m"=C[,2]-C[,4], "w-m"=C[,3]-C[,4])
# summarize these
pairwise_diffs_summary = posterior_summary(pairwise_diffs)We can think about these differences in the context of our type M and S errors. All of the group differences (save for boy-girl) have values of about 10 cm (or more), which is a meaningful (and noticeable) difference in human height. In addition, the credible intervals around these differences are small relative to their magnitudes. From this we can conclude that these differences are unlikely to really be the opposite sign (a type S error). In addition, since the group differences (save for boy-girl) are mostly large and the credible intervals do not go anywhere near zero, these differences seem unlikely to actually be some small and insignificant value near zero (a type S error).
But what about the boy-girl difference? The 95% credible interval of the difference between groups overlaps with zero. Does this mean there is no difference between the groups, i.e. \(\mu_{boy}=\mu_{girl}\) and \(\mu_{boy}-\mu_{girl}=0\)? No, it doesn’t. It just means that 0 is a value that is not so implausible for the difference between groups. The most probable difference is still 2.5. Consider a replication that found a group difference with a mean and 95% credible interval of 2.6 and [0.1, 5.1] respectively, rather than our values of 2.5 and [-0.8, 5.7]. Would such a difference constitute a successful replication or an opposing finding? If we focus on whether intervals cross zero to decide if effects are ‘real’, we might decide that these experiments do not agree: We found no difference in our first experiment but the replication did find a difference. In contrast, if we focus on the sign and magnitude of the effects and the uncertainty around their estimates, the two experiments have extremely compatible results and constitute a successful replication.
# summary of difference between boy and girl means
pairwise_diffs_summary[1,]
## Estimate Est.Error Q2.5 Q97.5
## 2.4643 1.6384 -0.7837 5.7199We might wonder, under what conditions can we accept that an effect or group difference is actually equal to zero. If the 95% (or some other high number) credible interval is relatively narrow and (more or less) centered around zero, the parameter value may very well be zero. It may also be a small number that has no practical effect on your dependent variable so that it doesn’t really matter what it is even if its not exactly equal to \(0.0000000 \dots\) . In either case, a coefficient with a narrow interval (roughly) centered at zero may not have an important role in the way you interpret your model in order to tell the ‘story’ of your data. However, it is extremely important to keep in mind that under a different parametrization of your model, or given a slightly different experimental design, it’s possible that an equivalent parameter may exhibit a clearly non-zero value.
Another thing to think about is, how large does an effect need to be before it matters, even if it is not zero? This is a similar approach to Kruschke’s Region of Practical Equivalence (ROPE, Kruschke 2018). Rather than worrying about whether things are exactly zero or not, Kruschke suggests we think about when things are so small that they may as well be zero. For human height, apparent and veridical, we define meaningful differences as those that 1) are likely to be different from zero, and 2) have magnitudes of at least around 1 cm (about 0.5 inches). We establish this lower limit simply based off the fact that people may describe themselves as 175 cm (or 5’10.5”) but rarely make distinctions smaller than that (e.g. people rarely distinguish 175 cm and 175.2 cm). In addition, governments, doctors, and even institutions like the NBA which are extremely interested in height will rarely measure to within less than 1 cm (half an inch). As a result, although a difference of 0.1 cm may very well be consistently different from zero, in the context of human height a difference of this magnitude has little to no practical significance in most situations.
In our case, we will conclude that the difference in apparent height between boys and girls may very well be ‘real’ and may reflect stereotypes that male speakers are generally taller than female speakers, whether or not this is actually true for speakers of this age. We might also suggest that if we want to know more about the precise value of this difference, future research might investigate this question with a larger number of speakers, listeners, or repetitions. By increasing the sample size, researchers would narrow the parameter likelihoods (discussed in section 2.7.3), thereby narrowing their posterior credible intervals (given new data that’s roughly the same as the data we have).
Finally, we want to directly address the fact that our discussion above regarding accepting effects as ‘real’ or as ‘zero’ may seem to be full of vagueness and equivocation. This is by design. Sometimes, researchers want to wash their hands of a difficult decision and let the model decide. However, the researcher cannot ‘pass the buck’ and let the model ‘prove’ that something is zero or non-zero. The model can certainly tell you that, given your data and model structure, something is likely to be zero or non zero. However, transferring that knowledge to the ‘real world’ is an epistemological or ontological claim (or both), and not a statistical one. To some extent, the statistical claim underlies the claim about the real world, but it is not enough on its own. To really make a claim about an effect being zero or not, the researcher requires statistical evidence in addition to domain knowledge about the real world. As a result, we can give general (vague) guidelines about situations when your models suggest something is zero or non zero, but we cannot say with certainty that in such and such a situation you can always make claims about the real world based solely on your statistical model.
For example, because of the relationship between sample size, likelihoods, and credible intervals, any arbitrarily small effect will be distinguishable from zero given a large enough sample. Imagine a situation where we have an effect of 0.1 cm that that has a 95% credible interval of [0.09, 0.11]. Our model is telling us that this effect is extremely unlikely to equal zero, and yet we are unlikely to care much about an effect this small. However, we can only know this by knowing something about human height and meaningful variation therein, and this is domain-specific knowledge that must come from the researcher interpreting the model.
7.8 Factors with more than two levels
Before moving on, we want to provide an example of how to recover interaction terms that are not estimated by your model when you have factors with more than two levels. This is a modest extension of the principles we have discussed so far, however, it’s useful to provide an explicit example. Below, we generate some toy data that includes two factors with four levels each (A, B), and an interaction between them (AB).
A = rep (factor(1:4), each = 60)
B = rep (factor(1:4), 60)
A_ = c(-2,1,2,-1)
B_ = c(4,2,-3,-3)
AB_ = matrix (c(-1,1,-1,1,
1,-1,1,-1,
-1,1,-1,1,
1,-1,1,-1),4,4)
error = rnorm(240)
y = A_[A] + B_[B] + AB_[cbind(A,B)] + error
toy_data = data.frame (y, A, B)We use this to fit a simple model with brms:
toy_model = brm (y ~ A*B, data = toy_data)
## Compiling Stan program...
## Start samplingAnd inspect the fixed effects:
fixef (toy_model)
## Estimate Est.Error Q2.5 Q97.5
## Intercept 0.02768 0.06593 -0.1049 0.1569
## A1 -2.10172 0.11882 -2.3394 -1.8672
## A2 1.09184 0.11453 0.8710 1.3232
## A3 1.92469 0.11882 1.6963 2.1542
## B1 4.02158 0.11775 3.7847 4.2484
## B2 2.08544 0.11764 1.8567 2.3178
## B3 -3.11586 0.11982 -3.3485 -2.8756
## A1:B1 -1.13546 0.20105 -1.5227 -0.7469
## A2:B1 1.22386 0.20184 0.8174 1.6255
## A3:B1 -1.20820 0.20252 -1.5987 -0.8064
## A1:B2 0.86307 0.19948 0.4691 1.2506
## A2:B2 -0.85335 0.20244 -1.2481 -0.4599
## A3:B2 0.75987 0.20433 0.3672 1.1688
## A1:B3 -0.77577 0.20288 -1.1653 -0.3664
## A2:B3 0.74222 0.20478 0.3327 1.1433
## A3:B3 -0.82375 0.20784 -1.2436 -0.4235As expected, we see that we estimate 3 of 4 factor levels for the \(A\) and \(B\) factors respectively. This means that we will be missing the fourth level of each factor. Below, we present factor levels in a grid, and indicate the missing level in parentheses.
\[ \begin{equation} \begin{split} \begin{bmatrix} A1 & A2 & A3 & (A4) \\ \end{bmatrix} \\\\ \begin{bmatrix} B1 & B2 & B3 & (B4) \\ \end{bmatrix} \end{split} \tag{7.13} \end{equation} \]
In order to recover the missing levels of \(A\) and \(B\) (under sum coding), we have to find the negative sum of the estimated levels. We place this value in the corresponding grid location seen below.
\[ \begin{equation} \begin{split} \begin{bmatrix} A1 & A2 & A3 & -(A1+A2+A3) \\ \end{bmatrix} \\\\ \begin{bmatrix} B1 & B2 & B3 & -(B1+B2+B3) \\ \end{bmatrix} \end{split} \tag{7.13} \end{equation} \]
As noted above, the number of interaction terms we can estimate will generally be equal to (the number of levels in \(A\) - 1)\(\times\)(the number of levels in \(B\) - 1). As a result, we know that we will only be able to estimate 9 out of the 16 total interaction terms. We have arranged these terms in a similar grid as seen above, and again placed the missing terms in parentheses. In the grid below, levels of \(A\) vary across columns and levels of \(B\) vary across rows.
\[ \begin{equation} \begin{bmatrix} A1 \colon B1 & A2 \colon B1 & A3 \colon B1 & (A4 \colon B1)\\ A1 \colon B2 & A2 \colon B2 & A3 \colon B2 & (A4 \colon B2)\\ A1 \colon B3 & A2 \colon B3 & A3 \colon B3 & (A4 \colon B3)\\ (A1 \colon B4) & (A2 \colon B4) & (A3 \colon B4) & (A4 \colon B4)\\ \end{bmatrix}\tag{7.14} \end{equation} \]
Recovering missing interaction values is no more complicated but can be a bit tedious for large numbers of missing interactions. The missing values at the end of each row are simply the negative sum of the elements present in the row, and the missing values at the end of each column are simply the negative sum of the elements present in the column. We show this for only one row and one column so that the matrix will fit on the page, but the same process can be applied to all rows and columns in the matrix.
\[ \begin{equation} \begin{bmatrix} A1 \colon B1 & A2 \colon B1 & A3 \colon B1 & -(A1 \colon B1+A2 \colon B1+A3 \colon B1) \\ A1 \colon B2 & A2 \colon B2 & A3 \colon B2 & (A4 \colon B2)\\ A1 \colon B3 & A2 \colon B3 & A3 \colon B3 & (A4 \colon B3)\\ -(A1 \colon B1+A1 \colon B2+A1 \colon B3) & (A2 \colon B4) & (A3 \colon B4) & (A4 \colon B4)\\ \end{bmatrix}\tag{7.14} \end{equation} \]
You can get the missing element in the bottom right corner above by following the same process once you have recovered the remaining elements from the final row or column. For example, the interaction in the bottom corner is the negative sum of the other three terms in the final column and in the final row.
\[ \begin{equation} \begin{split} A4 \colon B4 = -(A4 \colon B1+A4 \colon B2+A4 \colon B3) \\ A4 \colon B4 = -(A1 \colon B4+A2 \colon B4+A3 \colon B4) \\ \end{split} \tag{7.14} \end{equation} \]
Of course, these values can all be recovered easily using the hypothesis (or short_hypothesis) function.
7.9 ‘Traditionalists’ corner
In traditionalists corner, we’re going to compare the output of brms to some more ‘traditional’ approaches. We’re not going to talk about the traditional models in any detail, the focus of this section is simply to highlight the similarities between different approaches, and to point out where to find equivalent information in the different models. If you are already familiar with these approaches, these sections may be helpful. If not, some of the information provided here may not make much sense, although it may still be helpful. If you want to know more about the statistical methods being discussed here, please see the preface for a list of suggested background reading in statistics.
7.9.1 Bayesian multilevel models vs. lmer
This is going to be a short one. One shortcoming when it comes to using lmer to fit models including factors with many levels is that there is no easy way to compare group effects solely based on the information contained in the model. For example, we can fit the model below which encodes the difference between each group mean and the overall mean.
lmer_four_groups = lme4::lmer (height ~ C + (C|L) + (1|S), data = exp_data)
summary (lmer_four_groups)$coefficients
## Estimate Std. Error t value
## (Intercept) 158.344 1.040 152.228
## C1 -9.543 1.300 -7.341
## C2 -11.126 1.332 -8.355
## C3 14.604 1.371 10.655The results provided by lmer are very similar to those provided by brm:
brms::fixef (model_four_groups)
## Estimate Est.Error Q2.5 Q97.5
## Intercept 158.581 1.083 156.41 160.685
## C1 -9.318 1.310 -11.87 -6.682
## C2 -12.082 1.494 -15.07 -9.110
## C3 15.286 1.327 12.59 17.950However, it is not as straightforward to test for differences between parameter values in our lmer models. The same could be said for the combination of parameters as is necessary for the investigation of group means and simple effects. The investigation of differences and combinations of parameters is definitely possible in lmer for many cases, however this will involve either the re-parametrization of the model or the use of additional packages that allow for these sorts of questions to be asked. As a result, the comparisons or transformations that can be carried out is limited by the functionalities of the packages that carry out these comparison This can be contrasted with the ease and flexibility with which we can compare parameter estimates using Bayesian models.
7.10 Exercises
The analyses in the main body of the text all involve only the unmodified ‘actual’ resonance level (in exp_data). Responses for the stimuli with the simulate ‘big’ resonance are reserved for exercises throughout. You can get the ‘big’ resonance in the exp_ex data frame, or all data in the exp_data_all data frame.
Fit and interpret one or more of the suggested models:
Easy: Analyze the (pre-fit) model that’s exactly like
model_interaction, except using the data inexp_ex(bmmb::get_model("7_model_interaction_ex.RDS")).Medium: Fit a model like
model_interaction, but replace either apparent gender or apparent age with the resonance factor.Hard: Fit a model including apparent age, apparent gender, and resonance. Include all interactions between these factors for both fixed and random effects.
In any case, describe the model, present and explain the results, and include some figures.
7.11 References
Gelman, A., Hill, J., & Yajima, M. (2012). Why we (usually) don’t have to worry about multiple comparisons. Journal of Research on Educational Effectiveness, 5(2), 189–211.
Gelman, A., & Carlin, J. (2014). Beyond power calculations: Assessing type S (sign) and type M (magnitude) errors. Perspectives on Psychological Science, 9(6), 641-651.
Gelman, A., Goodrich, B., Gabry, J., & Vehtari, A. (2019). R-squared for Bayesian regression models. The American Statistician.
Kruschke, J. K. (2018). Rejecting or accepting parameter values in Bayesian estimation. Advances in methods and practices in psychological science, 1(2), 270-280.
7.12 Plot Code
################################################################################
### Figure 7.1
################################################################################
knitr::include_graphics("_main_files/figure-html/Figure 7.1.jpg")
################################################################################
### Figure 7.2
################################################################################
par (mfrow = c(1,2), mar = c(4,4,1,1))
boxplot (height ~ C, data = exp_data, col = bmmb::cols[2:5],ylim=c(100,200),
xlab = "Apparent Category", ylab = "Apparent Height (cm)")
boxplot (height ~ C_v, data = exp_data, col = bmmb::cols[2:5],ylim=c(100,200),
xlab = "Veridical Category", ylab = "Apparent Height (cm)")
################################################################################
### Figure 7.3
################################################################################
par (mfrow = c(1,2), mar = c(2.5,2,1,1), oma = c(0,2,0,0))
all_groups = bmmb::short_hypothesis (model_four_groups,
c("C1 = 0","C2 = 0","C3 = 0", "-(C1+C2+C3)=0"))
brmplot (all_groups, col=bmmb::cols[2:5],ylim=c(-16,19),
labels = c("b","g","m","w"))
points (means - mean(means), pch=4,lwd=2, col=bmmb::cols[2:5],cex=3)
brmplot (comparisons, col=bmmb::cols[13:12],labels = c("b-g","m-w"))
mtext (side = 2, outer = TRUE, "Centimeters",adj = .6, cex=1.2, line = 0.8)
################################################################################
### Figure 7.4
################################################################################
knitr::include_graphics("_main_files/figure-html/Figure 7.4.jpg")
################################################################################
### Figure 7.5
################################################################################
par (mfrow = c(1,3), mar = c(3,2.5,3,1), oma = c(.1,2,.1,.1))
boxplot (height ~ G+A, data=exp_data, main = "Overall", ylab="Apparent height (cm)",
ylim = c(90,200),col=bmmb::cols[c(5,4,3,2)],
names = c("w","m","g","b"),xlab="",cex.lab=1.3,cex.axis=1.3)
mtext (side=2,"Apparent height (cm)", line = 3, cex = 0.9)
boxplot (height ~ G, data=exp_data, main="Gender", ylim =c(90,200),
col=bmmb::cols[7:8], names = c("female","male"),xlab="",ylab="",
cex.lab=1.3,cex.axis=1.3)
boxplot (height ~ A, data=exp_data, main="Adultness", ylim = c(90,200),
col=bmmb::cols[1:2], names = c("adult","child"),xlab="",ylab="",
cex.lab=1.3, cex.axis=1.3)
################################################################################
### Figure 7.6
################################################################################
par (mfrow = c(1,3), mar = c(2.5,4,3,1))
boxplot (exp_data$height ~ exp_data$C, ylim = c(110,200),
col = bmmb::cols[2:5], main = "Original data",xlab = "",
ylab = "Apparent Height (cm)", outline = FALSE)
boxplot (y_hat_no_re[,1] ~ exp_data$C, ylim = c(110,200), col = bmmb::cols[2:5],
main = "Prediction: No RE",xlab = "", ylab="", outline = FALSE)
boxplot (y_hat[,1] ~ exp_data$C, ylim = c(110,200), col = bmmb::cols[2:5],
main = "Prediction: With RE",xlab = "", ylab="", outline = FALSE)
################################################################################
### Figure 7.7
################################################################################
par (mfrow = c(1,4), mar = c(3,0.5,2,0.5), oma = c(0,4,0,0))
plot (0:1,tapply (exp_data$height,exp_data$G,mean), ylim = c(140,180),xaxt='n',
lwd=10,type='b',pch=16,cex=3,main="Gender",xlim=c(-.2,1.2),
ylab='',xlab='',col = bmmb::cols[1])
axis (at=0:1, labels = c('female','male'), side=1,cex.axis=1.3)
grid()
G = exp_data$G
A = exp_data$A
mtext ("Apparent Height (cm)", side = 2, outer = TRUE, line = 2.75, cex = 0.9)
bmmb::interactionplot (G, A, exp_data$height,lwd = 10, leg.y = 170,
leg.x=1.8,type='b',pch=c(16,16),cex=3, ylim = c(140,180),lty=c(1,1),
main="Gender:Age",ylab='', xlim = c(.8,2.2),
legend =FALSE,yaxt="n",xlab="",cex.axis=1.3,
col = c(bmmb::cols[12],bmmb::cols[10]),xaxt='n')
grid()
axis (at=1:2, labels = c('female','male'), side=1,cex.axis=1.3)
plot (0:1,tapply (exp_data$height,exp_data$A,mean), ylim=c(140,180),xaxt='n',lwd=10,
type='b',pch=16,cex=3, main = "Age",xlim=c(-.2,1.2),ylab='',xlab='',yaxt="n",
col = bmmb::cols[8])
axis (at=0:1, labels = c('adult','child'),side=1,cex.axis=1.3)
grid()
bmmb::interactionplot (A, G, exp_data$height,lwd = 10, leg.y = 170,
leg.x=1.8,type='b',pch=c(16,16),cex=3, ylim = c(140,180),lty=c(1,1),
main="Age:Gender",ylab='',cex.axis=1.3 ,
xlim = c(.8,2.2),legend=FALSE,yaxt="n",xlab="",
col = c(bmmb::cols[11],bmmb::cols[9]),xaxt='n')
axis (at=1:2, labels = c('adult','child'),side=1,cex.axis=1.3)
grid()
################################################################################
### Figure 7.8
################################################################################
par (mfrow = c(1,3), mar = c(3,0.5,3,0.5), oma = c(0,4,0,0))
bmmb::interactionplot (exp_data$A,exp_data$G, exp_data$height,lwd = 10, leg.y = 170,
leg.x=1.8,type='b',pch=c(16,16),cex=3, ylim = c(140,180),lty=c(1,1),
main="Real Data",ylab='', xlim = c(.8,2.2),legend=FALSE,
xlab="", col = c(bmmb::cols[8],bmmb::cols[9]),xaxt='n')
grid()
axis (at=1:2, labels = c('adult','child'),side=1,cex.axis=1.3)
mtext ("Apparent Height (cm)", side = 2, outer = TRUE, line = 2.75, cex = 0.9)
bmmb::interactionplot (exp_data$A,exp_data$G, y_post_pred_no_re[,1],lwd = 10, leg.y = 170,
leg.x=1.8,type='b',pch=c(16,16),cex=3, ylim = c(140,180),lty=c(1,1),
main="Post. Pred: No RE",ylab='', xlim = c(.8,2.2),
legend =FALSE,yaxt="n",xlab="",xaxt='n',
col = c(bmmb::cols[4],bmmb::cols[3]))
grid()
axis (at=1:2, labels = c('adult','child'),side=1,cex.axis=1.3)
bmmb::interactionplot (exp_data$A,exp_data$G, y_post_pred[,1],lwd = 10, leg.y = 170,
leg.x=1.8,type='b',pch=c(16,16),cex=3, ylim = c(140,180),lty=c(1,1),
main="Post. Pred: With RE",ylab='', xlim = c(.8,2.2),legend=FALSE,
yaxt="n",xlab="", col = c(bmmb::cols[15],bmmb::cols[14]),xaxt='n')
grid()
axis (at=1:2, labels = c('adult','child'),side=1,cex.axis=1.3)
################################################################################
### Figure 7.9
###############################################################################
knitr::include_graphics("_main_files/figure-html/Figure 7.9.jpg")
###############################################################################
### Figure 7.10
###############################################################################
ps = aggregate (exp_data$height~ exp_data$G+ exp_data$A, FUN=mean)[,3]
par (mfrow = c(1,4), mar = c(2.5,0.2,2.5,0.2), oma = c(0,4,0,0))
#layout (mat = t(c(1,2,3,4)), widths = c(.28,.28,.28,.16))
interactionplot (exp_data$G, exp_data$A, y_post_pred_no_re[,1],
col=bmmb::cols[7], lwd=10,xaxt ='n',
type = 'l', pch = c(16,16), cex = 2, ylim = c(140,180),xlab="",
main="No Interaction, no RE", legend = FALSE,lty=1,
ylab = 'Apparent Height (cm)', xlim = c(.97,2.03),cex.main=1.1)
segments (1,ps[1],2,ps[2], col = bmmb::cols[8],lty=1,lwd=4)
segments (1,ps[3],2,ps[4], col = bmmb::cols[8],lty=1,lwd=4)
grid()
mtext ("Apparent Height (cm)", side = 2, outer = TRUE, line = 2.5, cex = 0.9)
axis (side=1,at=1:2,c("Female","Male"),cex.axis=1.2)
interactionplot (exp_data$G, exp_data$A, y_post_pred[,1],
col=bmmb::cols[7], lwd=10,lty=1,xaxt ='n',
type = 'l', pch = c(16,16), cex = 2, ylim = c(140,180),xlab="",
main="No Interaction, with RE", legend = FALSE,yaxt="n",
ylab = '', xlim = c(.97,2.03),cex.main=1.1)
segments (1,ps[1],2,ps[2], col = bmmb::cols[8],lty=1,lwd=4)
segments (1,ps[3],2,ps[4], col = bmmb::cols[8],lty=1,lwd=4)
grid()
axis (side=1,at=1:2,c("Female","Male"),cex.axis=1.2)
interactionplot (exp_data$G, exp_data$A, y_post_pred_no_re_int[,1],
col=bmmb::cols[7], lwd=10,xaxt ='n',
type = 'l', pch = c(16,16), cex = 2, ylim = c(140,180),xlab="",
main="Interaction, no RE", legend = FALSE,yaxt="n",lty=1,
ylab = '', xlim = c(.97,2.03),cex.main=1.1)
segments (1,ps[1],2,ps[2], col = bmmb::cols[8],lty=1,lwd=4)
segments (1,ps[3],2,ps[4], col = bmmb::cols[8],lty=1,lwd=4)
grid()
axis (side=1,at=1:2,c("Female","Male"),cex.axis=1.2)
interactionplot (exp_data$G, exp_data$A, y_post_pred_int[,1],
col=bmmb::cols[7], lwd=10,lty=1,xaxt ='n',
type = 'l', pch = c(16,16), cex = 2, ylim = c(140,180),xlab="",
main="Interaction, with RE", legend = FALSE, yaxt="n",
ylab = '', xlim = c(.97,2.03),cex.main=1.1)
segments (1,ps[1],2,ps[2], col = bmmb::cols[8],lty=1,lwd=4)
segments (1,ps[3],2,ps[4], col = bmmb::cols[8],lty=1,lwd=4)
grid()
axis (side=1,at=1:2,c("Female","Male"),cex.axis=1.2)
legend (1.25,165,legend = c("Observed", "Predicted"),bty='n',
lwd=4,col=bmmb::cols[7:8], cex=1.2)
###############################################################################
### Figure 7.11
##############################################################################
C = attributes(means_pred_interaction)$samples
pairwise_diffs = cbind(C[,1]-C[,2],C[,1]-C[,3],C[,1]-C[,4],
C[,2]-C[,3],C[,2]-C[,4],C[,3]-C[,4])
pairwise_diffs_summary = posterior_summary(pairwise_diffs)
par (mfrow = c(1,2), mar = c(4,2,1,1), oma = c(0,2,0,0))
layout (mat = t(c(1,2)), widths= c(.4,.6))
brmplot (fixef(model_interaction)[-1,])
brmplot (pairwise_diffs_summary,labels = c("b-g","b-w","b-m","g-w","g-m","w-m"))
mtext (side = 2, outer = TRUE, "Centimeters",adj = .6, cex=1.2, line = 0.8)